Note
Click here to download the full example code
Permutation t-test on source data with spatio-temporal clustering¶
This example tests if the evoked response is significantly different between two conditions across subjects. Here just for demonstration purposes we simulate data from multiple subjects using one subject’s data. The multiple comparisons problem is addressed with a cluster-level permutation test across space and time.
# Authors: Alexandre Gramfort <alexandre.gramfort@inria.fr>
# Eric Larson <larson.eric.d@gmail.com>
# License: BSD (3-clause)
import os.path as op
import numpy as np
from numpy.random import randn
from scipy import stats as stats
import mne
from mne.epochs import equalize_epoch_counts
from mne.stats import (spatio_temporal_cluster_1samp_test,
summarize_clusters_stc)
from mne.minimum_norm import apply_inverse, read_inverse_operator
from mne.datasets import sample
print(__doc__)
Set parameters¶
data_path = sample.data_path()
raw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
event_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw-eve.fif'
subjects_dir = data_path + '/subjects'
src_fname = subjects_dir + '/fsaverage/bem/fsaverage-ico-5-src.fif'
tmin = -0.2
tmax = 0.3 # Use a lower tmax to reduce multiple comparisons
# Setup for reading the raw data
raw = mne.io.read_raw_fif(raw_fname)
events = mne.read_events(event_fname)
Out:
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Read epochs for all channels, removing a bad one¶
raw.info['bads'] += ['MEG 2443']
picks = mne.pick_types(raw.info, meg=True, eog=True, exclude='bads')
event_id = 1 # L auditory
reject = dict(grad=1000e-13, mag=4000e-15, eog=150e-6)
epochs1 = mne.Epochs(raw, events, event_id, tmin, tmax, picks=picks,
baseline=(None, 0), reject=reject, preload=True)
event_id = 3 # L visual
epochs2 = mne.Epochs(raw, events, event_id, tmin, tmax, picks=picks,
baseline=(None, 0), reject=reject, preload=True)
# Equalize trial counts to eliminate bias (which would otherwise be
# introduced by the abs() performed below)
equalize_epoch_counts([epochs1, epochs2])
Out:
Not setting metadata
Not setting metadata
72 matching events found
Setting baseline interval to [-0.19979521315838786, 0.0] sec
Applying baseline correction (mode: mean)
Created an SSP operator (subspace dimension = 3)
4 projection items activated
Loading data for 72 events and 76 original time points ...
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on MAG : ['MEG 1711']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
9 bad epochs dropped
Not setting metadata
Not setting metadata
73 matching events found
Setting baseline interval to [-0.19979521315838786, 0.0] sec
Applying baseline correction (mode: mean)
Created an SSP operator (subspace dimension = 3)
4 projection items activated
Loading data for 73 events and 76 original time points ...
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on GRAD : ['MEG 1333', 'MEG 1342']
Rejecting epoch based on EOG : ['EOG 061']
6 bad epochs dropped
Dropped 0 epochs:
Dropped 4 epochs: 10, 25, 26, 40
Transform to source space¶
fname_inv = data_path + '/MEG/sample/sample_audvis-meg-oct-6-meg-inv.fif'
snr = 3.0
lambda2 = 1.0 / snr ** 2
method = "dSPM" # use dSPM method (could also be MNE, sLORETA, or eLORETA)
inverse_operator = read_inverse_operator(fname_inv)
sample_vertices = [s['vertno'] for s in inverse_operator['src']]
# Let's average and compute inverse, resampling to speed things up
evoked1 = epochs1.average()
evoked1.resample(50, npad='auto')
condition1 = apply_inverse(evoked1, inverse_operator, lambda2, method)
evoked2 = epochs2.average()
evoked2.resample(50, npad='auto')
condition2 = apply_inverse(evoked2, inverse_operator, lambda2, method)
# Let's only deal with t > 0, cropping to reduce multiple comparisons
condition1.crop(0, None)
condition2.crop(0, None)
tmin = condition1.tmin
tstep = condition1.tstep * 1000 # convert to milliseconds
Out:
Reading inverse operator decomposition from /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis-meg-oct-6-meg-inv.fif...
Reading inverse operator info...
[done]
Reading inverse operator decomposition...
[done]
305 x 305 full covariance (kind = 1) found.
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Noise covariance matrix read.
22494 x 22494 diagonal covariance (kind = 2) found.
Source covariance matrix read.
22494 x 22494 diagonal covariance (kind = 6) found.
Orientation priors read.
22494 x 22494 diagonal covariance (kind = 5) found.
Depth priors read.
Did not find the desired covariance matrix (kind = 3)
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
2 source spaces read
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Source spaces transformed to the inverse solution coordinate frame
Removing projector <Projection | Average EEG reference, active : True, n_channels : 60>
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 63
Created the regularized inverter
Created an SSP operator (subspace dimension = 3)
Created the whitener using a noise covariance matrix with rank 302 (3 small eigenvalues omitted)
Computing noise-normalization factors (dSPM)...
[done]
Applying inverse operator to "1"...
Picked 305 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Computing residual...
Explained 69.4% variance
Combining the current components...
dSPM...
[done]
Removing projector <Projection | Average EEG reference, active : True, n_channels : 60>
Preparing the inverse operator for use...
Scaled noise and source covariance from nave = 1 to nave = 63
Created the regularized inverter
Created an SSP operator (subspace dimension = 3)
Created the whitener using a noise covariance matrix with rank 302 (3 small eigenvalues omitted)
Computing noise-normalization factors (dSPM)...
[done]
Applying inverse operator to "3"...
Picked 305 channels from the data
Computing inverse...
Eigenleads need to be weighted ...
Computing residual...
Explained 70.7% variance
Combining the current components...
dSPM...
[done]
Transform to common cortical space¶
Normally you would read in estimates across several subjects and morph them to the same cortical space (e.g. fsaverage). For example purposes, we will simulate this by just having each “subject” have the same response (just noisy in source space) here.
Note
Note that for 7 subjects with a two-sided statistical test, the minimum significance under a permutation test is only p = 1/(2 ** 6) = 0.015, which is large.
n_vertices_sample, n_times = condition1.data.shape
n_subjects = 7
print('Simulating data for %d subjects.' % n_subjects)
# Let's make sure our results replicate, so set the seed.
np.random.seed(0)
X = randn(n_vertices_sample, n_times, n_subjects, 2) * 10
X[:, :, :, 0] += condition1.data[:, :, np.newaxis]
X[:, :, :, 1] += condition2.data[:, :, np.newaxis]
Out:
Simulating data for 7 subjects.
It’s a good idea to spatially smooth the data, and for visualization purposes, let’s morph these to fsaverage, which is a grade 5 source space with vertices 0:10242 for each hemisphere. Usually you’d have to morph each subject’s data separately (and you might want to use morph_data instead), but here since all estimates are on ‘sample’ we can use one morph matrix for all the heavy lifting.
# Read the source space we are morphing to
src = mne.read_source_spaces(src_fname)
fsave_vertices = [s['vertno'] for s in src]
morph_mat = mne.compute_source_morph(
src=inverse_operator['src'], subject_to='fsaverage',
spacing=fsave_vertices, subjects_dir=subjects_dir).morph_mat
n_vertices_fsave = morph_mat.shape[0]
# We have to change the shape for the dot() to work properly
X = X.reshape(n_vertices_sample, n_times * n_subjects * 2)
print('Morphing data.')
X = morph_mat.dot(X) # morph_mat is a sparse matrix
X = X.reshape(n_vertices_fsave, n_times, n_subjects, 2)
Out:
Reading a source space...
[done]
Reading a source space...
[done]
2 source spaces read
Morphing data.
Finally, we want to compare the overall activity levels in each condition, the diff is taken along the last axis (condition). The negative sign makes it so condition1 > condition2 shows up as “red blobs” (instead of blue).
Compute statistic¶
To use an algorithm optimized for spatio-temporal clustering, we just pass the spatial adjacency matrix (instead of spatio-temporal)
print('Computing adjacency.')
adjacency = mne.spatial_src_adjacency(src)
# Note that X needs to be a multi-dimensional array of shape
# samples (subjects) x time x space, so we permute dimensions
X = np.transpose(X, [2, 1, 0])
# Now let's actually do the clustering. This can take a long time...
# Here we set the threshold quite high to reduce computation.
p_threshold = 0.001
t_threshold = -stats.distributions.t.ppf(p_threshold / 2., n_subjects - 1)
print('Clustering.')
T_obs, clusters, cluster_p_values, H0 = clu = \
spatio_temporal_cluster_1samp_test(X, adjacency=adjacency, n_jobs=1,
threshold=t_threshold, buffer_size=None,
verbose=True)
# Now select the clusters that are sig. at p < 0.05 (note that this value
# is multiple-comparisons corrected).
good_cluster_inds = np.where(cluster_p_values < 0.05)[0]
Out:
Computing adjacency.
-- number of adjacent vertices : 20484
Clustering.
stat_fun(H1): min=-25.828905 max=32.550985
Running initial clustering
Found 345 clusters
Permuting 63 times (exact test)...
0%| | : 0/63 [00:00<?, ?it/s]
2%|1 | : 1/63 [00:00<00:02, 25.27it/s]
3%|3 | : 2/63 [00:00<00:02, 27.29it/s]
6%|6 | : 4/63 [00:00<00:01, 37.88it/s]
8%|7 | : 5/63 [00:00<00:01, 35.72it/s]
10%|9 | : 6/63 [00:00<00:01, 34.38it/s]
13%|#2 | : 8/63 [00:00<00:01, 38.93it/s]
14%|#4 | : 9/63 [00:00<00:01, 37.40it/s]
17%|#7 | : 11/63 [00:00<00:01, 40.57it/s]
19%|#9 | : 12/63 [00:00<00:01, 39.10it/s]
22%|##2 | : 14/63 [00:00<00:01, 41.57it/s]
24%|##3 | : 15/63 [00:00<00:01, 40.20it/s]
25%|##5 | : 16/63 [00:00<00:01, 39.05it/s]
29%|##8 | : 18/63 [00:00<00:01, 41.09it/s]
30%|### | : 19/63 [00:00<00:01, 39.98it/s]
33%|###3 | : 21/63 [00:00<00:01, 41.74it/s]
35%|###4 | : 22/63 [00:00<00:01, 40.65it/s]
38%|###8 | : 24/63 [00:00<00:00, 42.23it/s]
40%|###9 | : 25/63 [00:00<00:00, 41.19it/s]
41%|####1 | : 26/63 [00:00<00:00, 40.25it/s]
44%|####4 | : 28/63 [00:00<00:00, 41.71it/s]
46%|####6 | : 29/63 [00:00<00:00, 40.79it/s]
49%|####9 | : 31/63 [00:00<00:00, 42.14it/s]
51%|##### | : 32/63 [00:00<00:00, 41.23it/s]
54%|#####3 | : 34/63 [00:00<00:00, 42.49it/s]
56%|#####5 | : 35/63 [00:00<00:00, 41.60it/s]
59%|#####8 | : 37/63 [00:00<00:00, 42.79it/s]
60%|###### | : 38/63 [00:00<00:00, 41.90it/s]
62%|######1 | : 39/63 [00:00<00:00, 41.10it/s]
65%|######5 | : 41/63 [00:00<00:00, 42.25it/s]
67%|######6 | : 42/63 [00:01<00:00, 41.44it/s]
70%|######9 | : 44/63 [00:01<00:00, 42.55it/s]
71%|#######1 | : 45/63 [00:01<00:00, 41.75it/s]
75%|#######4 | : 47/63 [00:01<00:00, 42.81it/s]
76%|#######6 | : 48/63 [00:01<00:00, 42.01it/s]
79%|#######9 | : 50/63 [00:01<00:00, 43.04it/s]
81%|######## | : 51/63 [00:01<00:00, 42.24it/s]
83%|########2 | : 52/63 [00:01<00:00, 41.50it/s]
86%|########5 | : 54/63 [00:01<00:00, 42.52it/s]
87%|########7 | : 55/63 [00:01<00:00, 41.78it/s]
90%|######### | : 57/63 [00:01<00:00, 42.77it/s]
92%|#########2| : 58/63 [00:01<00:00, 42.02it/s]
95%|#########5| : 60/63 [00:01<00:00, 42.98it/s]
97%|#########6| : 61/63 [00:01<00:00, 42.23it/s]
98%|#########8| : 62/63 [00:01<00:00, 41.52it/s]
100%|##########| : 63/63 [00:01<00:00, 42.12it/s]
Computing cluster p-values
Done.
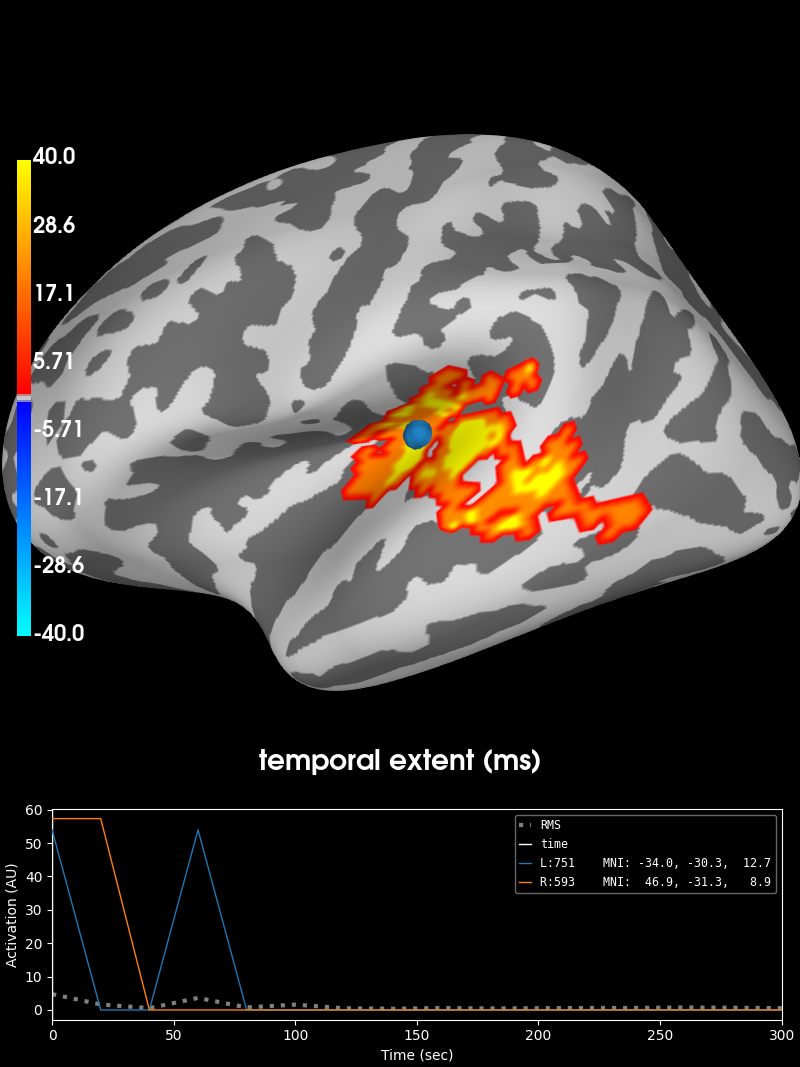
Visualize the clusters¶
print('Visualizing clusters.')
# Now let's build a convenient representation of each cluster, where each
# cluster becomes a "time point" in the SourceEstimate
stc_all_cluster_vis = summarize_clusters_stc(clu, tstep=tstep,
vertices=fsave_vertices,
subject='fsaverage')
# Let's actually plot the first "time point" in the SourceEstimate, which
# shows all the clusters, weighted by duration.
subjects_dir = op.join(data_path, 'subjects')
# blue blobs are for condition A < condition B, red for A > B
brain = stc_all_cluster_vis.plot(
hemi='both', views='lateral', subjects_dir=subjects_dir,
time_label='temporal extent (ms)', size=(800, 800),
smoothing_steps=5, clim=dict(kind='value', pos_lims=[0, 1, 40]))
# brain.save_image('clusters.png')
Out:
Visualizing clusters.
Total running time of the script: ( 0 minutes 19.646 seconds)
Estimated memory usage: 113 MB