Note
Click here to download the full example code
Whitening evoked data with a noise covariance¶
Evoked data are loaded and then whitened using a given noise covariance matrix. It’s an excellent quality check to see if baseline signals match the assumption of Gaussian white noise during the baseline period.
Covariance estimation and diagnostic plots are based on 1.
References¶
- 1
Denis A. Engemann and Alexandre Gramfort. Automated model selection in covariance estimation and spatial whitening of MEG and EEG signals. NeuroImage, 108:328–342, 2015. doi:10.1016/j.neuroimage.2014.12.040.
# Authors: Alexandre Gramfort <alexandre.gramfort@inria.fr>
# Denis A. Engemann <denis.engemann@gmail.com>
#
# License: BSD (3-clause)
import mne
from mne import io
from mne.datasets import sample
from mne.cov import compute_covariance
print(__doc__)
Set parameters
data_path = sample.data_path()
raw_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw.fif'
event_fname = data_path + '/MEG/sample/sample_audvis_filt-0-40_raw-eve.fif'
raw = io.read_raw_fif(raw_fname, preload=True)
raw.filter(1, 40, n_jobs=1, fir_design='firwin')
raw.info['bads'] += ['MEG 2443'] # bads + 1 more
events = mne.read_events(event_fname)
# let's look at rare events, button presses
event_id, tmin, tmax = 2, -0.2, 0.5
reject = dict(mag=4e-12, grad=4000e-13, eeg=80e-6)
epochs = mne.Epochs(raw, events, event_id, tmin, tmax, picks=('meg', 'eeg'),
baseline=None, reject=reject, preload=True)
# Uncomment next line to use fewer samples and study regularization effects
# epochs = epochs[:20] # For your data, use as many samples as you can!
Out:
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Reading 0 ... 41699 = 0.000 ... 277.709 secs...
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 1 - 40 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 1.00
- Lower transition bandwidth: 1.00 Hz (-6 dB cutoff frequency: 0.50 Hz)
- Upper passband edge: 40.00 Hz
- Upper transition bandwidth: 10.00 Hz (-6 dB cutoff frequency: 45.00 Hz)
- Filter length: 497 samples (3.310 sec)
Not setting metadata
Not setting metadata
73 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 4)
4 projection items activated
Loading data for 73 events and 106 original time points ...
Rejecting epoch based on EEG : ['EEG 001', 'EEG 002', 'EEG 003', 'EEG 007']
Rejecting epoch based on EEG : ['EEG 001', 'EEG 002', 'EEG 003', 'EEG 004', 'EEG 005', 'EEG 006', 'EEG 007']
Rejecting epoch based on EEG : ['EEG 003', 'EEG 007']
Rejecting epoch based on EEG : ['EEG 001', 'EEG 002', 'EEG 003', 'EEG 006', 'EEG 007']
Rejecting epoch based on MAG : ['MEG 1711']
Rejecting epoch based on EEG : ['EEG 007']
Rejecting epoch based on EEG : ['EEG 008', 'EEG 009']
Rejecting epoch based on EEG : ['EEG 001', 'EEG 002', 'EEG 003', 'EEG 007']
Rejecting epoch based on EEG : ['EEG 001', 'EEG 002', 'EEG 003', 'EEG 006', 'EEG 007']
Rejecting epoch based on EEG : ['EEG 001', 'EEG 002', 'EEG 003', 'EEG 007']
Rejecting epoch based on EEG : ['EEG 001', 'EEG 002', 'EEG 003', 'EEG 007']
Rejecting epoch based on EEG : ['EEG 001', 'EEG 007']
12 bad epochs dropped
Compute covariance using automated regularization
method_params = dict(diagonal_fixed=dict(mag=0.01, grad=0.01, eeg=0.01))
noise_covs = compute_covariance(epochs, tmin=None, tmax=0, method='auto',
return_estimators=True, verbose=True, n_jobs=1,
projs=None, rank=None,
method_params=method_params)
# With "return_estimator=True" all estimated covariances sorted
# by log-likelihood are returned.
print('Covariance estimates sorted from best to worst')
for c in noise_covs:
print("%s : %s" % (c['method'], c['loglik']))
Out:
Computing rank from data with rank=None
Using tolerance 3.8e-09 (2.2e-16 eps * 305 dim * 5.6e+04 max singular value)
Estimated rank (mag + grad): 302
MEG: rank 302 computed from 305 data channels with 3 projectors
Using tolerance 1.4e-11 (2.2e-16 eps * 59 dim * 1.1e+03 max singular value)
Estimated rank (eeg): 58
EEG: rank 58 computed from 59 data channels with 1 projector
Created an SSP operator (subspace dimension = 4)
Setting small MEG eigenvalues to zero (without PCA)
Setting small EEG eigenvalues to zero (without PCA)
Reducing data rank from 364 -> 360
Estimating covariance using SHRUNK
Done.
Estimating covariance using DIAGONAL_FIXED
MAG regularization : 0.01
GRAD regularization : 0.01
EEG regularization : 0.01
Done.
Estimating covariance using EMPIRICAL
Done.
Using cross-validation to select the best estimator.
MAG regularization : 0.01
GRAD regularization : 0.01
EEG regularization : 0.01
MAG regularization : 0.01
GRAD regularization : 0.01
EEG regularization : 0.01
MAG regularization : 0.01
GRAD regularization : 0.01
EEG regularization : 0.01
Number of samples used : 1891
log-likelihood on unseen data (descending order):
shrunk: -1628.686
diagonal_fixed: -1659.785
empirical: -1802.103
[done]
Covariance estimates sorted from best to worst
shrunk : -1628.6862795945235
diagonal_fixed : -1659.7854492948447
empirical : -1802.1031535820846
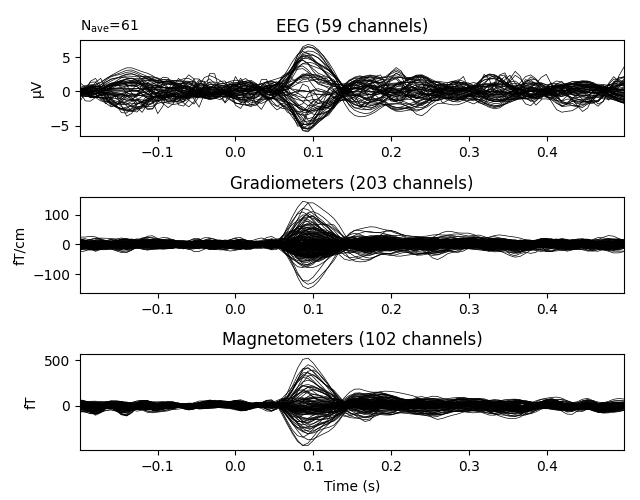
Show the evoked data:
evoked = epochs.average()
evoked.plot(time_unit='s') # plot evoked response
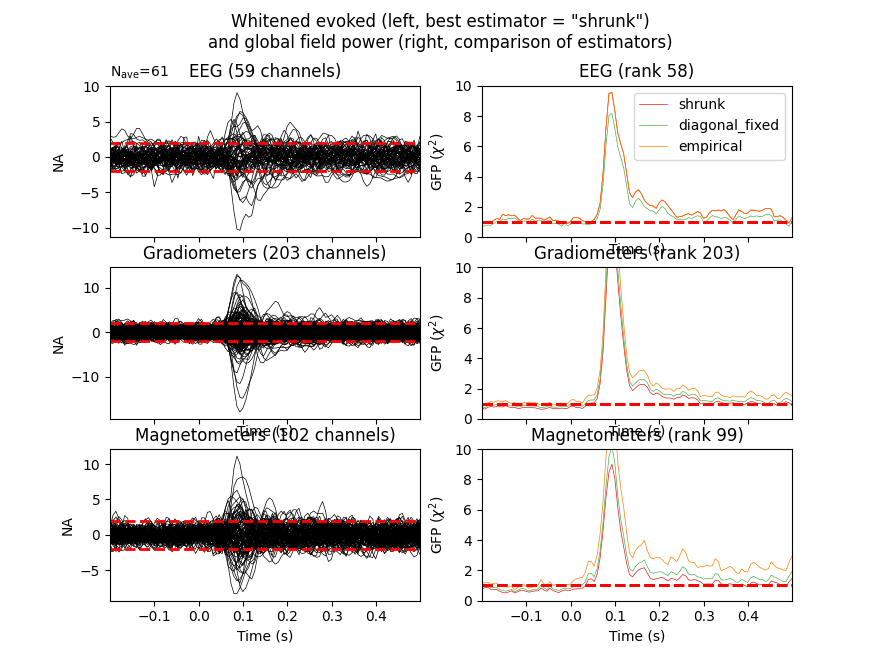
We can then show whitening for our various noise covariance estimates.
Here we should look to see if baseline signals match the assumption of Gaussian white noise. we expect values centered at 0 within 2 standard deviations for 95% of the time points.
For the Global field power we expect a value of 1.
evoked.plot_white(noise_covs, time_unit='s')
Out:
Computing rank from covariance with rank=None
Using tolerance 8.2e-14 (2.2e-16 eps * 59 dim * 6.3 max singular value)
Estimated rank (eeg): 58
EEG: rank 58 computed from 59 data channels with 1 projector
Computing rank from covariance with rank=None
Using tolerance 2.8e-13 (2.2e-16 eps * 203 dim * 6.2 max singular value)
Estimated rank (grad): 203
GRAD: rank 203 computed from 203 data channels with 0 projectors
Computing rank from covariance with rank=None
Using tolerance 3.6e-14 (2.2e-16 eps * 102 dim * 1.6 max singular value)
Estimated rank (mag): 99
MAG: rank 99 computed from 102 data channels with 3 projectors
Computing rank from covariance with rank=None
Using tolerance 8.2e-14 (2.2e-16 eps * 59 dim * 6.3 max singular value)
Estimated rank (eeg): 58
EEG: rank 58 computed from 59 data channels with 1 projector
Computing rank from covariance with rank=None
Using tolerance 2.8e-13 (2.2e-16 eps * 203 dim * 6.3 max singular value)
Estimated rank (grad): 203
GRAD: rank 203 computed from 203 data channels with 0 projectors
Computing rank from covariance with rank=None
Using tolerance 3.7e-14 (2.2e-16 eps * 102 dim * 1.6 max singular value)
Estimated rank (mag): 99
MAG: rank 99 computed from 102 data channels with 3 projectors
Computing rank from covariance with rank=None
Using tolerance 8.2e-14 (2.2e-16 eps * 59 dim * 6.3 max singular value)
Estimated rank (eeg): 58
EEG: rank 58 computed from 59 data channels with 1 projector
Computing rank from covariance with rank=None
Using tolerance 2.8e-13 (2.2e-16 eps * 203 dim * 6.3 max singular value)
Estimated rank (grad): 203
GRAD: rank 203 computed from 203 data channels with 0 projectors
Computing rank from covariance with rank=None
Using tolerance 3.7e-14 (2.2e-16 eps * 102 dim * 1.6 max singular value)
Estimated rank (mag): 99
MAG: rank 99 computed from 102 data channels with 3 projectors
Created an SSP operator (subspace dimension = 4)
Computing rank from covariance with rank={'eeg': 58, 'grad': 203, 'mag': 99, 'meg': 302}
Setting small MEG eigenvalues to zero (without PCA)
Setting small EEG eigenvalues to zero (without PCA)
Created the whitener using a noise covariance matrix with rank 360 (4 small eigenvalues omitted)
Created an SSP operator (subspace dimension = 4)
Computing rank from covariance with rank={'eeg': 58, 'grad': 203, 'mag': 99, 'meg': 302}
Setting small MEG eigenvalues to zero (without PCA)
Setting small EEG eigenvalues to zero (without PCA)
Created the whitener using a noise covariance matrix with rank 360 (4 small eigenvalues omitted)
Created an SSP operator (subspace dimension = 4)
Computing rank from covariance with rank={'eeg': 58, 'grad': 203, 'mag': 99, 'meg': 302}
Setting small MEG eigenvalues to zero (without PCA)
Setting small EEG eigenvalues to zero (without PCA)
Created the whitener using a noise covariance matrix with rank 360 (4 small eigenvalues omitted)
Total running time of the script: ( 0 minutes 18.511 seconds)
Estimated memory usage: 128 MB