Note
Go to the end to download the full example code
XDAWN Decoding From EEG data#
ERP decoding with Xdawn [1][2]. For each event type, a set of spatial Xdawn filters are trained and applied on the signal. Channels are concatenated and rescaled to create features vectors that will be fed into a logistic regression.
# Authors: Alexandre Barachant <alexandre.barachant@gmail.com>
#
# License: BSD-3-Clause
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import StratifiedKFold
from sklearn.pipeline import make_pipeline
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import classification_report, confusion_matrix
from sklearn.preprocessing import MinMaxScaler
from mne import io, pick_types, read_events, Epochs, EvokedArray, create_info
from mne.datasets import sample
from mne.preprocessing import Xdawn
from mne.decoding import Vectorizer
print(__doc__)
data_path = sample.data_path()
Set parameters and read data
meg_path = data_path / "MEG" / "sample"
raw_fname = meg_path / "sample_audvis_filt-0-40_raw.fif"
event_fname = meg_path / "sample_audvis_filt-0-40_raw-eve.fif"
tmin, tmax = -0.1, 0.3
event_id = {
"Auditory/Left": 1,
"Auditory/Right": 2,
"Visual/Left": 3,
"Visual/Right": 4,
}
n_filter = 3
# Setup for reading the raw data
raw = io.read_raw_fif(raw_fname, preload=True)
raw.filter(1, 20, fir_design="firwin")
events = read_events(event_fname)
picks = pick_types(raw.info, meg=False, eeg=True, stim=False, eog=False, exclude="bads")
epochs = Epochs(
raw,
events,
event_id,
tmin,
tmax,
proj=False,
picks=picks,
baseline=None,
preload=True,
verbose=False,
)
# Create classification pipeline
clf = make_pipeline(
Xdawn(n_components=n_filter),
Vectorizer(),
MinMaxScaler(),
LogisticRegression(penalty="l1", solver="liblinear", multi_class="auto"),
)
# Get the labels
labels = epochs.events[:, -1]
# Cross validator
cv = StratifiedKFold(n_splits=10, shuffle=True, random_state=42)
# Do cross-validation
preds = np.empty(len(labels))
for train, test in cv.split(epochs, labels):
clf.fit(epochs[train], labels[train])
preds[test] = clf.predict(epochs[test])
# Classification report
target_names = ["aud_l", "aud_r", "vis_l", "vis_r"]
report = classification_report(labels, preds, target_names=target_names)
print(report)
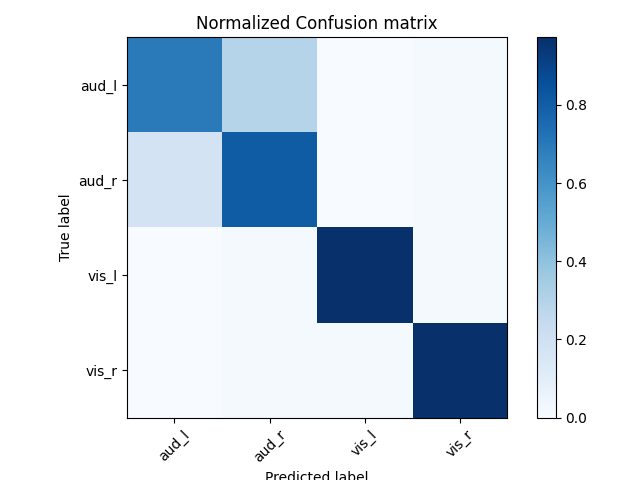
# Normalized confusion matrix
cm = confusion_matrix(labels, preds)
cm_normalized = cm.astype(float) / cm.sum(axis=1)[:, np.newaxis]
# Plot confusion matrix
fig, ax = plt.subplots(1)
im = ax.imshow(cm_normalized, interpolation="nearest", cmap=plt.cm.Blues)
ax.set(title="Normalized Confusion matrix")
fig.colorbar(im)
tick_marks = np.arange(len(target_names))
plt.xticks(tick_marks, target_names, rotation=45)
plt.yticks(tick_marks, target_names)
fig.tight_layout()
ax.set(ylabel="True label", xlabel="Predicted label")
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Reading 0 ... 41699 = 0.000 ... 277.709 secs...
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 1 - 20 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 1.00
- Lower transition bandwidth: 1.00 Hz (-6 dB cutoff frequency: 0.50 Hz)
- Upper passband edge: 20.00 Hz
- Upper transition bandwidth: 5.00 Hz (-6 dB cutoff frequency: 22.50 Hz)
- Filter length: 497 samples (3.310 s)
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 2 out of 2 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 4 out of 4 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 366 out of 366 | elapsed: 0.6s finished
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank='full'
EEG: rank 59 from info
Created an SSP operator (subspace dimension = 1)
Reducing data rank from 59 -> 59
Estimating covariance using EMPIRICAL
Done.
precision recall f1-score support
aud_l 0.79 0.69 0.74 72
aud_r 0.72 0.81 0.76 73
vis_l 0.99 0.97 0.98 73
vis_r 0.96 0.97 0.96 70
accuracy 0.86 288
macro avg 0.86 0.86 0.86 288
weighted avg 0.86 0.86 0.86 288
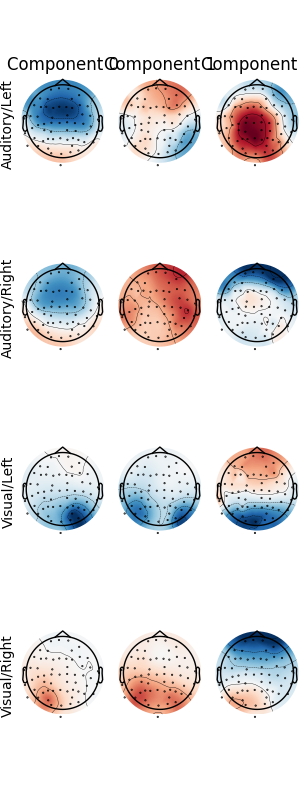
The patterns_ attribute of a fitted Xdawn instance (here from the last
cross-validation fold) can be used for visualization.
fig, axes = plt.subplots(
nrows=len(event_id), ncols=n_filter, figsize=(n_filter, len(event_id) * 2)
)
fitted_xdawn = clf.steps[0][1]
info = create_info(epochs.ch_names, 1, epochs.get_channel_types())
info.set_montage(epochs.get_montage())
for ii, cur_class in enumerate(sorted(event_id)):
cur_patterns = fitted_xdawn.patterns_[cur_class]
pattern_evoked = EvokedArray(cur_patterns[:n_filter].T, info, tmin=0)
pattern_evoked.plot_topomap(
times=np.arange(n_filter),
time_format="Component %d" if ii == 0 else "",
colorbar=False,
show_names=False,
axes=axes[ii],
show=False,
)
axes[ii, 0].set(ylabel=cur_class)
fig.tight_layout(h_pad=1.0, w_pad=1.0, pad=0.1)
References#
Total running time of the script: ( 0 minutes 7.402 seconds)
Estimated memory usage: 129 MB