Note
Go to the end to download the full example code
Shifting time-scale in evoked data#
# Author: Mainak Jas <mainak@neuro.hut.fi>
#
# License: BSD-3-Clause
import matplotlib.pyplot as plt
import mne
from mne.viz import tight_layout
from mne.datasets import sample
print(__doc__)
data_path = sample.data_path()
meg_path = data_path / "MEG" / "sample"
fname = meg_path / "sample_audvis-ave.fif"
# Reading evoked data
condition = "Left Auditory"
evoked = mne.read_evokeds(fname, condition=condition, baseline=(None, 0), proj=True)
ch_names = evoked.info["ch_names"]
picks = mne.pick_channels(ch_names=ch_names, include=["MEG 2332"])
# Create subplots
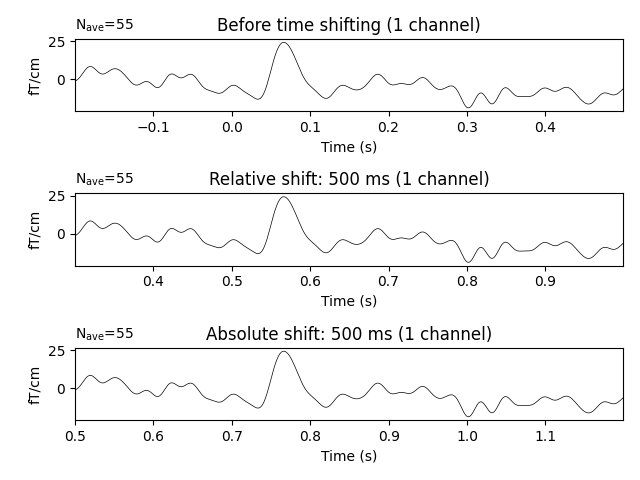
f, (ax1, ax2, ax3) = plt.subplots(3)
evoked.plot(
exclude=[],
picks=picks,
axes=ax1,
titles=dict(grad="Before time shifting"),
time_unit="s",
)
# Apply relative time-shift of 500 ms
evoked.shift_time(0.5, relative=True)
evoked.plot(
exclude=[],
picks=picks,
axes=ax2,
titles=dict(grad="Relative shift: 500 ms"),
time_unit="s",
)
# Apply absolute time-shift of 500 ms
evoked.shift_time(0.5, relative=False)
evoked.plot(
exclude=[],
picks=picks,
axes=ax3,
titles=dict(grad="Absolute shift: 500 ms"),
time_unit="s",
)
tight_layout()
Reading /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis-ave.fif ...
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Found the data of interest:
t = -199.80 ... 499.49 ms (Left Auditory)
0 CTF compensation matrices available
nave = 55 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
Applying baseline correction (mode: mean)
Need more than one channel to make topography for grad. Disabling interactivity.
Need more than one channel to make topography for grad. Disabling interactivity.
Need more than one channel to make topography for grad. Disabling interactivity.
Total running time of the script: ( 0 minutes 2.559 seconds)
Estimated memory usage: 10 MB