Note
Go to the end to download the full example code
Plotting topographic arrowmaps of evoked data#
Load evoked data and plot arrowmaps along with the topomap for selected time points. An arrowmap is based upon the Hosaka-Cohen transformation and represents an estimation of the current flow underneath the MEG sensors. They are a poor man’s MNE.
See [1] for details.
References#
# Authors: Sheraz Khan <sheraz@khansheraz.com>
#
# License: BSD-3-Clause
import numpy as np
import mne
from mne.datasets import sample
from mne.datasets.brainstorm import bst_raw
from mne import read_evokeds
from mne.viz import plot_arrowmap
print(__doc__)
path = sample.data_path()
fname = path / "MEG" / "sample" / "sample_audvis-ave.fif"
# load evoked data
condition = "Left Auditory"
evoked = read_evokeds(fname, condition=condition, baseline=(None, 0))
evoked_mag = evoked.copy().pick_types(meg="mag")
evoked_grad = evoked.copy().pick_types(meg="grad")
Reading /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis-ave.fif ...
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Found the data of interest:
t = -199.80 ... 499.49 ms (Left Auditory)
0 CTF compensation matrices available
nave = 55 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
Applying baseline correction (mode: mean)
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
Removing projector <Projection | Average EEG reference, active : True, n_channels : 60>
NOTE: pick_types() is a legacy function. New code should use inst.pick(...).
Removing projector <Projection | PCA-v1, active : True, n_channels : 102>
Removing projector <Projection | PCA-v2, active : True, n_channels : 102>
Removing projector <Projection | PCA-v3, active : True, n_channels : 102>
Removing projector <Projection | Average EEG reference, active : True, n_channels : 60>
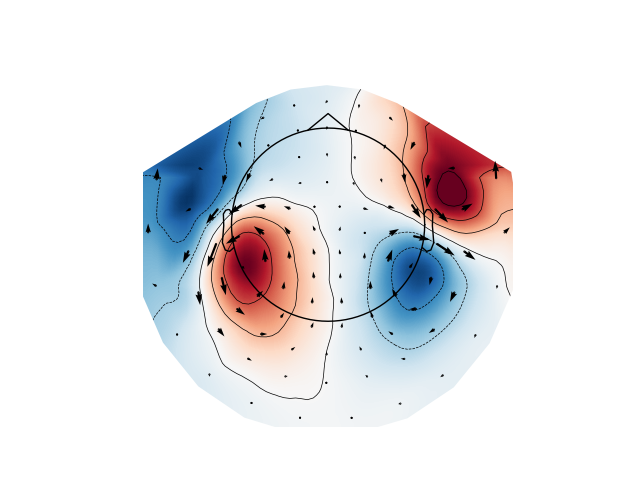
Plot magnetometer data as an arrowmap along with the topoplot at the time of the maximum sensor space activity:
max_time_idx = np.abs(evoked_mag.data).mean(axis=0).argmax()
plot_arrowmap(evoked_mag.data[:, max_time_idx], evoked_mag.info)
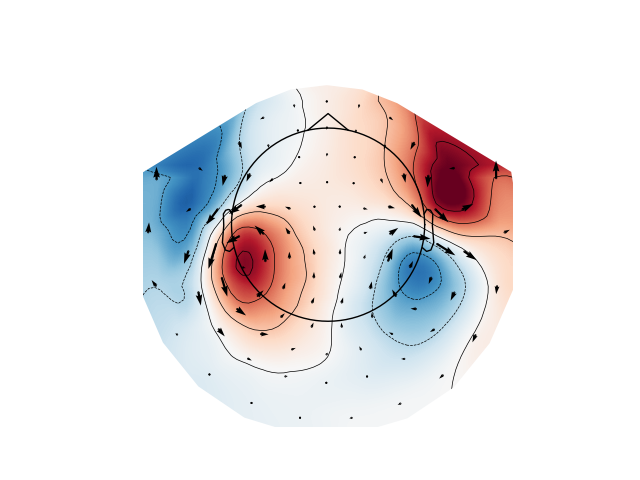
# Since planar gradiometers takes gradients along latitude and longitude,
# they need to be projected to the flatten manifold span by magnetometer
# or radial gradiometers before taking the gradients in the 2D Cartesian
# coordinate system for visualization on the 2D topoplot. You can use the
# ``info_from`` and ``info_to`` parameters to interpolate from
# gradiometer data to magnetometer data.
Plot gradiometer data as an arrowmap along with the topoplot at the time of the maximum sensor space activity:
plot_arrowmap(
evoked_grad.data[:, max_time_idx],
info_from=evoked_grad.info,
info_to=evoked_mag.info,
)
Computing dot products for 203 MEG channels...
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 2.1s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 2.1s finished
Computing cross products for 203 → 102 MEG channels...
Preparing the mapping matrix...
Truncating at 79/203 components to omit less than 0.0001 (9.2e-05)
Since Vectorview 102 system perform sparse spatial sampling of the magnetic field, data from the Vectorview (info_from) can be projected to the high density CTF 272 system (info_to) for visualization
Plot gradiometer data as an arrowmap along with the topoplot at the time of the maximum sensor space activity:
path = bst_raw.data_path()
raw_fname = path / "MEG" / "bst_raw" / "subj001_somatosensory_20111109_01_AUX-f.ds"
raw_ctf = mne.io.read_raw_ctf(raw_fname)
raw_ctf_info = mne.pick_info(
raw_ctf.info, mne.pick_types(raw_ctf.info, meg=True, ref_meg=False)
)
plot_arrowmap(
evoked_grad.data[:, max_time_idx],
info_from=evoked_grad.info,
info_to=raw_ctf_info,
scale=6e-10,
)
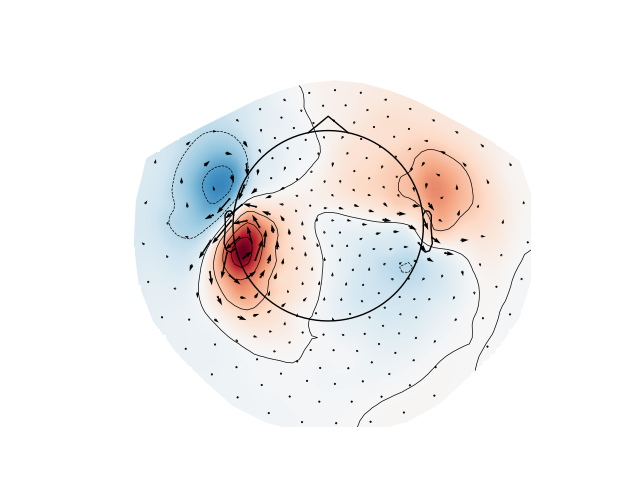
ds directory : /home/circleci/mne_data/MNE-brainstorm-data/bst_raw/MEG/bst_raw/subj001_somatosensory_20111109_01_AUX-f.ds
res4 data read.
hc data read.
Separate EEG position data file not present.
Quaternion matching (desired vs. transformed):
0.84 69.49 0.00 mm <-> 0.84 69.49 -0.00 mm (orig : -44.30 51.45 -252.43 mm) diff = 0.000 mm
-0.84 -69.49 0.00 mm <-> -0.84 -69.49 -0.00 mm (orig : 46.28 -53.58 -243.47 mm) diff = 0.000 mm
86.41 0.00 0.00 mm <-> 86.41 0.00 0.00 mm (orig : 63.60 55.82 -230.26 mm) diff = 0.000 mm
Coordinate transformations established.
Reading digitizer points from ['/home/circleci/mne_data/MNE-brainstorm-data/bst_raw/MEG/bst_raw/subj001_somatosensory_20111109_01_AUX-f.ds/subj00111092011.pos']...
Polhemus data for 3 HPI coils added
Device coordinate locations for 3 HPI coils added
Picked positions of 2 EEG channels from channel info
2 EEG locations added to Polhemus data.
Measurement info composed.
Finding samples for /home/circleci/mne_data/MNE-brainstorm-data/bst_raw/MEG/bst_raw/subj001_somatosensory_20111109_01_AUX-f.ds/subj001_somatosensory_20111109_01_AUX-f.meg4:
System clock channel is available, checking which samples are valid.
240 x 1800 = 432000 samples from 302 chs
Current compensation grade : 3
Removing 5 compensators from info because not all compensation channels were picked.
Computing dot products for 203 MEG channels...
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 2.1s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 2.1s finished
Computing cross products for 203 → 272 MEG channels...
Preparing the mapping matrix...
Truncating at 79/203 components to omit less than 0.0001 (9.2e-05)
Total running time of the script: ( 0 minutes 22.700 seconds)
Estimated memory usage: 16 MB