Tutorials#
These tutorials provide narrative explanations, sample code, and expected output for the most common MNE-Python analysis tasks. The emphasis here is on thorough explanations that get you up to speed quickly, at the expense of covering only a limited number of topics. The sections and tutorials are arranged in a fixed order, so in theory a new user should be able to progress through in order without encountering any cases where background knowledge is assumed and unexplained. More experienced users (i.e., those with significant experience analyzing EEG/MEG signals with different software) can probably skip around to just the topics they need without too much trouble.
Note
If tutorial-scripts contain plots and are run locally, using the
interactive flag with python -i tutorial_script.py
keeps them open.
Introductory tutorials#
These tutorials cover the basic EEG/MEG pipeline for event-related analysis,
introduce the mne.Info, events, and mne.Annotations
data structures, discuss how sensor locations are handled, and introduce some
of the configuration options available.
Reading data for different recording systems#
These tutorials cover the basics of loading EEG/MEG data into MNE-Python for various recording devices.
Working with CTF data: the Brainstorm auditory dataset
Working with continuous data#
These tutorials cover the basics of loading EEG/MEG data into MNE-Python, and
how to query, manipulate, annotate, plot, and export continuous data in the
Raw format.
Preprocessing#
These tutorials cover various preprocessing techniques for continuous data, as well as some diagnostic plotting methods.

Signal-space separation (SSS) and Maxwell filtering
Preprocessing functional near-infrared spectroscopy (fNIRS) data
Preprocessing optically pumped magnetometer (OPM) MEG data
Segmenting continuous data into epochs#
These tutorials cover epoched data, and how it differs from working with continuous data.
Estimating evoked responses#
These tutorials cover estimates of evoked responses (i.e., averages across several repetitions of an experimental condition).
Time-frequency analysis#
These tutorials cover frequency and time-frequency analysis of neural signals.

The Spectrum and EpochsSpectrum classes: frequency-domain data

Frequency-tagging: Basic analysis of an SSVEP/vSSR dataset
Forward models and source spaces#
These tutorials cover how the cortical source locations (source spaces) and forward models (AKA leadfield matrices) are defined.
Source localization and inverses#
These tutorials cover estimation of cortical activity from sensor recordings.
Source localization with equivalent current dipole (ECD) fit
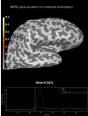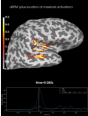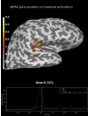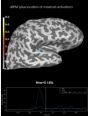
Source localization with MNE, dSPM, sLORETA, and eLORETA
The role of dipole orientations in distributed source localization
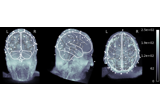
EEG source localization given electrode locations on an MRI
Statistical analysis of sensor data#
These tutorials describe some approaches to statistical analysis of sensor-level data.
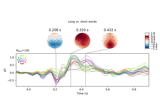
Visualising statistical significance thresholds on EEG data
Non-parametric 1 sample cluster statistic on single trial power
Non-parametric between conditions cluster statistic on single trial power

Mass-univariate twoway repeated measures ANOVA on single trial power

Spatiotemporal permutation F-test on full sensor data
Statistical analysis of source estimates#
These tutorials cover within-subject statistical analysis of source estimates.
Permutation t-test on source data with spatio-temporal clustering
2 samples permutation test on source data with spatio-temporal clustering
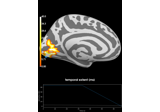
Repeated measures ANOVA on source data with spatio-temporal clustering
Machine learning models of neural activity#
These tutorials cover some of the machine learning methods available in MNE-Python.
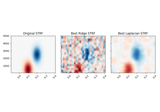
Spectro-temporal receptive field (STRF) estimation on continuous data
Clinical applications#
These tutorials illustrate some clinical use cases.
MNE-GUI-addons examples#
The mne_gui_addons package supports some clinical use cases:
MNE-Python examples#
MNE-Python also supports some clinical use cases directly:
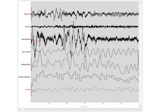
Sleep stage classification from polysomnography (PSG) data
Simulation#
These tutorials describe how to populate MNE-Python data structures with arbitrary data, using the array-based constructors and the simulation submodule.