Note
Go to the end to download the full example code
Temporal whitening with AR model#
Here we fit an AR model to the data and use it to temporally whiten the signals.
# Authors: Alexandre Gramfort <alexandre.gramfort@inria.fr>
#
# License: BSD-3-Clause
import numpy as np
from scipy import signal
import matplotlib.pyplot as plt
import mne
from mne.time_frequency import fit_iir_model_raw
from mne.datasets import sample
print(__doc__)
data_path = sample.data_path()
meg_path = data_path / "MEG" / "sample"
raw_fname = meg_path / "sample_audvis_raw.fif"
proj_fname = meg_path / "sample_audvis_ecg-proj.fif"
raw = mne.io.read_raw_fif(raw_fname)
proj = mne.read_proj(proj_fname)
raw.add_proj(proj)
raw.info["bads"] = ["MEG 2443", "EEG 053"] # mark bad channels
# Set up pick list: Gradiometers - bad channels
picks = mne.pick_types(raw.info, meg="grad", exclude="bads")
order = 5 # define model order
picks = picks[:1]
# Estimate AR models on raw data
b, a = fit_iir_model_raw(raw, order=order, picks=picks, tmin=60, tmax=180)
d, times = raw[0, 10000:20000] # look at one channel from now on
d = d.ravel() # make flat vector
innovation = signal.convolve(d, a, "valid")
d_ = signal.lfilter(b, a, innovation) # regenerate the signal
d_ = np.r_[d_[0] * np.ones(order), d_] # dummy samples to keep signal length
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_raw.fif...
Read a total of 3 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Range : 25800 ... 192599 = 42.956 ... 320.670 secs
Ready.
Read a total of 6 projection items:
ECG-planar-999--0.200-0.400-PCA-01 (1 x 203) idle
ECG-planar-999--0.200-0.400-PCA-02 (1 x 203) idle
ECG-axial-999--0.200-0.400-PCA-01 (1 x 102) idle
ECG-axial-999--0.200-0.400-PCA-02 (1 x 102) idle
ECG-eeg-999--0.200-0.400-PCA-01 (1 x 59) idle
ECG-eeg-999--0.200-0.400-PCA-02 (1 x 59) idle
6 projection items deactivated
Plot the different time series and PSDs
plt.close("all")
plt.figure()
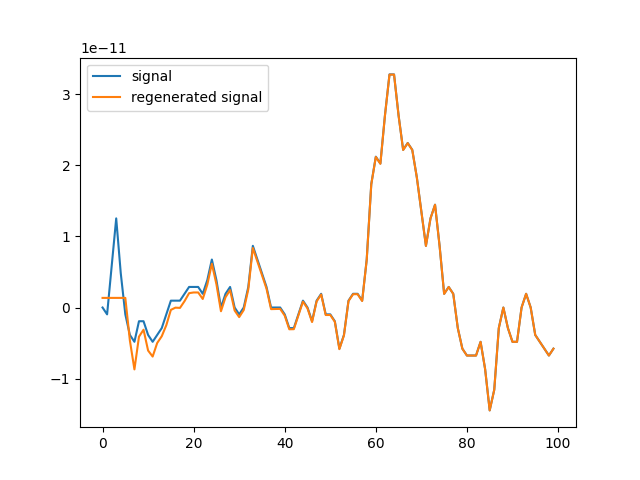
plt.plot(d[:100], label="signal")
plt.plot(d_[:100], label="regenerated signal")
plt.legend()
plt.figure()
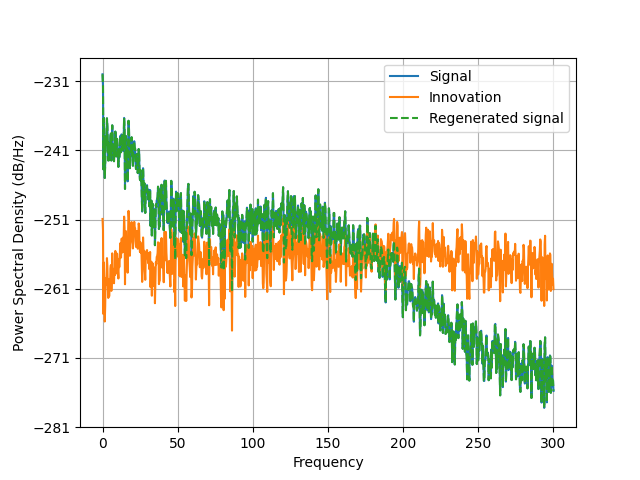
plt.psd(d, Fs=raw.info["sfreq"], NFFT=2048)
plt.psd(innovation, Fs=raw.info["sfreq"], NFFT=2048)
plt.psd(d_, Fs=raw.info["sfreq"], NFFT=2048, linestyle="--")
plt.legend(("Signal", "Innovation", "Regenerated signal"))
plt.show()
Total running time of the script: ( 0 minutes 4.278 seconds)
Estimated memory usage: 10 MB