Note
Go to the end to download the full example code
Reduce EOG artifacts through regression#
Reduce artifacts by regressing the EOG channels onto the rest of the channels and then subtracting the EOG signal.
This is a quick example to show the most basic application of the technique. See the tutorial for a more thorough explanation that demonstrates more advanced approaches.
# Author: Marijn van Vliet <w.m.vanvliet@gmail.com>
#
# License: BSD (3-clause)
Import packages and load data#
We begin as always by importing the necessary Python modules and loading some data, in this case the MNE sample dataset.
import mne
from mne.datasets import sample
from mne.preprocessing import EOGRegression
from matplotlib import pyplot as plt
print(__doc__)
data_path = sample.data_path()
raw_fname = data_path / "MEG" / "sample" / "sample_audvis_filt-0-40_raw.fif"
# Read raw data
raw = mne.io.read_raw_fif(raw_fname, preload=True)
events = mne.find_events(raw, "STI 014")
# Highpass filter to eliminate slow drifts
raw.filter(0.3, None, picks="all")
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Reading 0 ... 41699 = 0.000 ... 277.709 secs...
319 events found
Event IDs: [ 1 2 3 4 5 32]
Filtering raw data in 1 contiguous segment
Setting up high-pass filter at 0.3 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal highpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 0.30
- Lower transition bandwidth: 0.30 Hz (-6 dB cutoff frequency: 0.15 Hz)
- Filter length: 1653 samples (11.009 s)
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 2 out of 2 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 3 out of 3 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 4 out of 4 | elapsed: 0.0s remaining: 0.0s
[Parallel(n_jobs=1)]: Done 376 out of 376 | elapsed: 0.9s finished
Perform regression and remove EOG#
# Fit the regression model
weights = EOGRegression().fit(raw)
raw_clean = weights.apply(raw, copy=True)
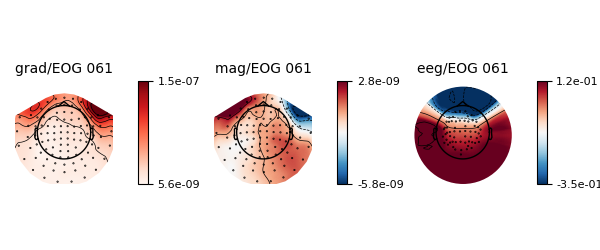
# Show the filter weights in a topomap
weights.plot()
Created an SSP operator (subspace dimension = 4)
4 projection items activated
SSP projectors applied...
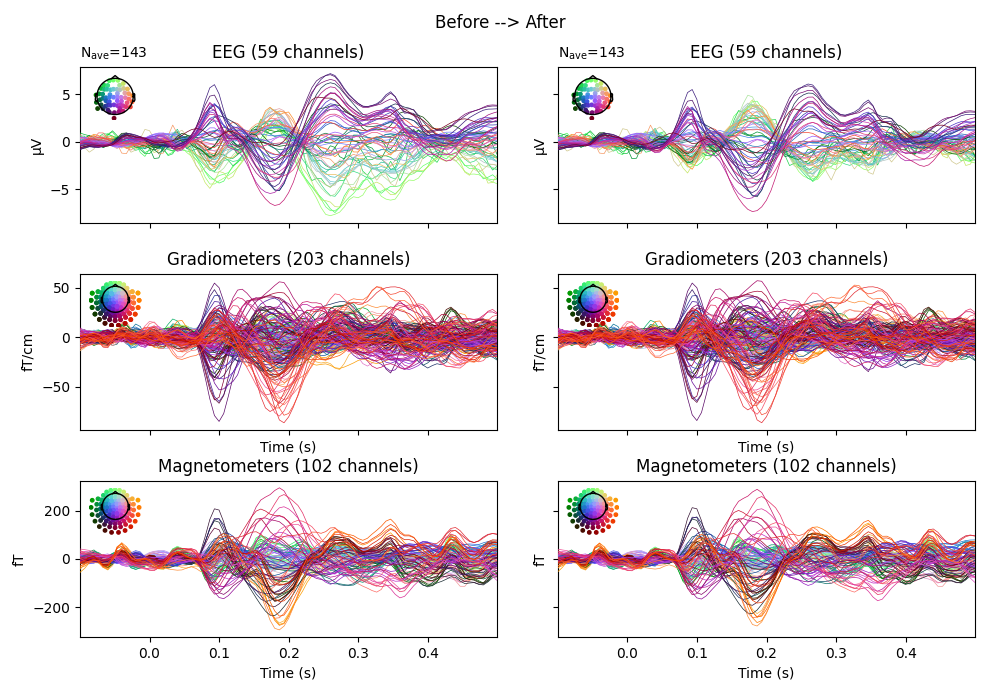
Before/after comparison#
Let’s compare the signal before and after cleaning with EOG regression. This is best visualized by extracting epochs and plotting the evoked potential.
tmin, tmax = -0.1, 0.5
event_id = {"visual/left": 3, "visual/right": 4}
evoked_before = mne.Epochs(
raw, events, event_id, tmin, tmax, baseline=(tmin, 0)
).average()
evoked_after = mne.Epochs(
raw_clean, events, event_id, tmin, tmax, baseline=(tmin, 0)
).average()
# Create epochs after EOG correction
epochs_after = mne.Epochs(raw_clean, events, event_id, tmin, tmax, baseline=(tmin, 0))
evoked_after = epochs_after.average()
fig, ax = plt.subplots(nrows=3, ncols=2, figsize=(10, 7), sharex=True, sharey="row")
evoked_before.plot(axes=ax[:, 0], spatial_colors=True)
evoked_after.plot(axes=ax[:, 1], spatial_colors=True)
fig.subplots_adjust(
top=0.905, bottom=0.09, left=0.08, right=0.975, hspace=0.325, wspace=0.145
)
fig.suptitle("Before --> After")
Not setting metadata
143 matching events found
Applying baseline correction (mode: mean)
Created an SSP operator (subspace dimension = 4)
4 projection items activated
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
Not setting metadata
143 matching events found
Applying baseline correction (mode: mean)
Created an SSP operator (subspace dimension = 4)
4 projection items activated
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
Not setting metadata
143 matching events found
Applying baseline correction (mode: mean)
Created an SSP operator (subspace dimension = 4)
4 projection items activated
NOTE: pick_channels() is a legacy function. New code should use inst.pick(...).
Total running time of the script: ( 0 minutes 11.330 seconds)
Estimated memory usage: 249 MB