Note
Click here to download the full example code
Motor imagery decoding from EEG data using the Common Spatial Pattern (CSP)¶
Decoding of motor imagery applied to EEG data decomposed using CSP. A classifier is then applied to features extracted on CSP-filtered signals.
See https://en.wikipedia.org/wiki/Common_spatial_pattern and 1. The EEGBCI dataset is documented in 2 and is available at PhysioNet 3.
# Authors: Martin Billinger <martin.billinger@tugraz.at>
#
# License: BSD-3-Clause
import numpy as np
import matplotlib.pyplot as plt
from sklearn.pipeline import Pipeline
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.model_selection import ShuffleSplit, cross_val_score
from mne import Epochs, pick_types, events_from_annotations
from mne.channels import make_standard_montage
from mne.io import concatenate_raws, read_raw_edf
from mne.datasets import eegbci
from mne.decoding import CSP
print(__doc__)
# #############################################################################
# # Set parameters and read data
# avoid classification of evoked responses by using epochs that start 1s after
# cue onset.
tmin, tmax = -1., 4.
event_id = dict(hands=2, feet=3)
subject = 1
runs = [6, 10, 14] # motor imagery: hands vs feet
raw_fnames = eegbci.load_data(subject, runs)
raw = concatenate_raws([read_raw_edf(f, preload=True) for f in raw_fnames])
eegbci.standardize(raw) # set channel names
montage = make_standard_montage('standard_1005')
raw.set_montage(montage)
# strip channel names of "." characters
raw.rename_channels(lambda x: x.strip('.'))
# Apply band-pass filter
raw.filter(7., 30., fir_design='firwin', skip_by_annotation='edge')
events, _ = events_from_annotations(raw, event_id=dict(T1=2, T2=3))
picks = pick_types(raw.info, meg=False, eeg=True, stim=False, eog=False,
exclude='bads')
# Read epochs (train will be done only between 1 and 2s)
# Testing will be done with a running classifier
epochs = Epochs(raw, events, event_id, tmin, tmax, proj=True, picks=picks,
baseline=None, preload=True)
epochs_train = epochs.copy().crop(tmin=1., tmax=2.)
labels = epochs.events[:, -1] - 2
Out:
Extracting EDF parameters from /home/circleci/mne_data/MNE-eegbci-data/files/eegmmidb/1.0.0/S001/S001R06.edf...
EDF file detected
Setting channel info structure...
Creating raw.info structure...
Reading 0 ... 19999 = 0.000 ... 124.994 secs...
Extracting EDF parameters from /home/circleci/mne_data/MNE-eegbci-data/files/eegmmidb/1.0.0/S001/S001R10.edf...
EDF file detected
Setting channel info structure...
Creating raw.info structure...
Reading 0 ... 19999 = 0.000 ... 124.994 secs...
Extracting EDF parameters from /home/circleci/mne_data/MNE-eegbci-data/files/eegmmidb/1.0.0/S001/S001R14.edf...
EDF file detected
Setting channel info structure...
Creating raw.info structure...
Reading 0 ... 19999 = 0.000 ... 124.994 secs...
Filtering raw data in 3 contiguous segments
Setting up band-pass filter from 7 - 30 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 7.00
- Lower transition bandwidth: 2.00 Hz (-6 dB cutoff frequency: 6.00 Hz)
- Upper passband edge: 30.00 Hz
- Upper transition bandwidth: 7.50 Hz (-6 dB cutoff frequency: 33.75 Hz)
- Filter length: 265 samples (1.656 sec)
Used Annotations descriptions: ['T1', 'T2']
Not setting metadata
Not setting metadata
45 matching events found
No baseline correction applied
0 projection items activated
Loading data for 45 events and 801 original time points ...
0 bad epochs dropped
Classification with linear discrimant analysis
# Define a monte-carlo cross-validation generator (reduce variance):
scores = []
epochs_data = epochs.get_data()
epochs_data_train = epochs_train.get_data()
cv = ShuffleSplit(10, test_size=0.2, random_state=42)
cv_split = cv.split(epochs_data_train)
# Assemble a classifier
lda = LinearDiscriminantAnalysis()
csp = CSP(n_components=4, reg=None, log=True, norm_trace=False)
# Use scikit-learn Pipeline with cross_val_score function
clf = Pipeline([('CSP', csp), ('LDA', lda)])
scores = cross_val_score(clf, epochs_data_train, labels, cv=cv, n_jobs=1)
# Printing the results
class_balance = np.mean(labels == labels[0])
class_balance = max(class_balance, 1. - class_balance)
print("Classification accuracy: %f / Chance level: %f" % (np.mean(scores),
class_balance))
# plot CSP patterns estimated on full data for visualization
csp.fit_transform(epochs_data, labels)
csp.plot_patterns(epochs.info, ch_type='eeg', units='Patterns (AU)', size=1.5)
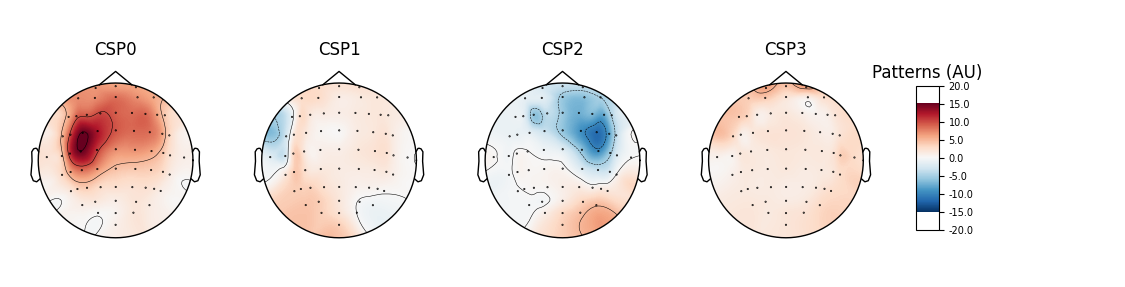
Out:
Computing rank from data with rank=None
Using tolerance 9.7e-05 (2.2e-16 eps * 64 dim * 6.8e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.3e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 9.5e-05 (2.2e-16 eps * 64 dim * 6.7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.2e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00012 (2.2e-16 eps * 64 dim * 8.6e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 9.6e-05 (2.2e-16 eps * 64 dim * 6.7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00011 (2.2e-16 eps * 64 dim * 7.5e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00011 (2.2e-16 eps * 64 dim * 7.6e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00012 (2.2e-16 eps * 64 dim * 8.1e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.2e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00012 (2.2e-16 eps * 64 dim * 8.5e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 9.5e-05 (2.2e-16 eps * 64 dim * 6.7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00012 (2.2e-16 eps * 64 dim * 8.6e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 9.6e-05 (2.2e-16 eps * 64 dim * 6.7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00012 (2.2e-16 eps * 64 dim * 8.3e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.2e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.1e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00011 (2.2e-16 eps * 64 dim * 7.7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.3e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Classification accuracy: 0.933333 / Chance level: 0.533333
Computing rank from data with rank=None
Using tolerance 0.00025 (2.2e-16 eps * 64 dim * 1.7e+10 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00026 (2.2e-16 eps * 64 dim * 1.9e+10 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Look at performance over time
sfreq = raw.info['sfreq']
w_length = int(sfreq * 0.5) # running classifier: window length
w_step = int(sfreq * 0.1) # running classifier: window step size
w_start = np.arange(0, epochs_data.shape[2] - w_length, w_step)
scores_windows = []
for train_idx, test_idx in cv_split:
y_train, y_test = labels[train_idx], labels[test_idx]
X_train = csp.fit_transform(epochs_data_train[train_idx], y_train)
X_test = csp.transform(epochs_data_train[test_idx])
# fit classifier
lda.fit(X_train, y_train)
# running classifier: test classifier on sliding window
score_this_window = []
for n in w_start:
X_test = csp.transform(epochs_data[test_idx][:, :, n:(n + w_length)])
score_this_window.append(lda.score(X_test, y_test))
scores_windows.append(score_this_window)
# Plot scores over time
w_times = (w_start + w_length / 2.) / sfreq + epochs.tmin
plt.figure()
plt.plot(w_times, np.mean(scores_windows, 0), label='Score')
plt.axvline(0, linestyle='--', color='k', label='Onset')
plt.axhline(0.5, linestyle='-', color='k', label='Chance')
plt.xlabel('time (s)')
plt.ylabel('classification accuracy')
plt.title('Classification score over time')
plt.legend(loc='lower right')
plt.show()
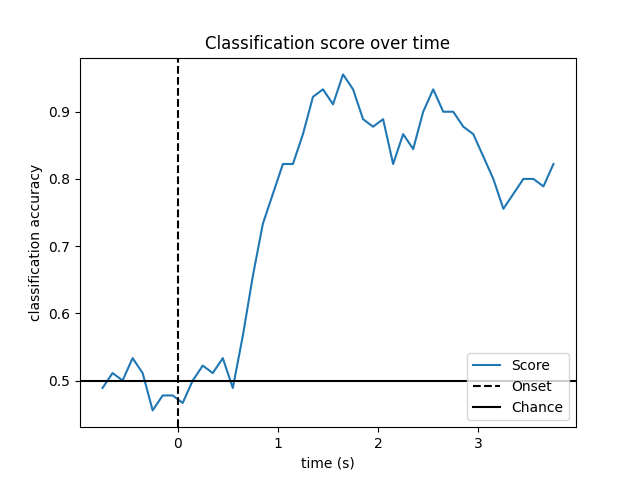
Out:
Computing rank from data with rank=None
Using tolerance 9.7e-05 (2.2e-16 eps * 64 dim * 6.8e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.3e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 9.5e-05 (2.2e-16 eps * 64 dim * 6.7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.2e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00012 (2.2e-16 eps * 64 dim * 8.6e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 9.6e-05 (2.2e-16 eps * 64 dim * 6.7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00011 (2.2e-16 eps * 64 dim * 7.5e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00011 (2.2e-16 eps * 64 dim * 7.6e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00012 (2.2e-16 eps * 64 dim * 8.1e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.2e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00012 (2.2e-16 eps * 64 dim * 8.5e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 9.5e-05 (2.2e-16 eps * 64 dim * 6.7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00012 (2.2e-16 eps * 64 dim * 8.6e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 9.6e-05 (2.2e-16 eps * 64 dim * 6.7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00012 (2.2e-16 eps * 64 dim * 8.3e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.2e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.1e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00011 (2.2e-16 eps * 64 dim * 7.7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.3e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
References¶
- 1
Zoltan J. Koles. The quantitative extraction and topographic mapping of the abnormal components in the clinical EEG. Electroencephalography and Clinical Neurophysiology, 79(6):440–447, 1991. doi:10.1016/0013-4694(91)90163-X.
- 2
Gerwin Schalk, Dennis J. McFarland, Thilo Hinterberger, Niels Birbaumer, and Jonathan R. Wolpaw. BCI2000: a general-purpose brain-computer interface (BCI) system. IEEE Transactions on Biomedical Engineering, 51(6):1034–1043, 2004. doi:10.1109/TBME.2004.827072.
- 3
Ary L. Goldberger, Luis A. N. Amaral, Leon Glass, Jeffrey M. Hausdorff, Plamen Ch. Ivanov, Roger G. Mark, Joseph E. Mietus, George B. Moody, Chung-Kang Peng, and H. Eugene Stanley. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation, 2000. doi:10.1161/01.CIR.101.23.e215.
Total running time of the script: ( 0 minutes 9.967 seconds)
Estimated memory usage: 9 MB