Note
Click here to download the full example code
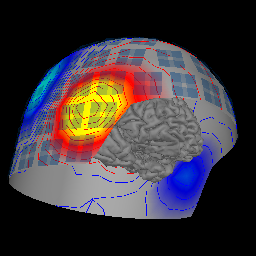
Plot the MNE brain and helmet¶
This tutorial shows how to make the MNE helmet + brain image.
Out:
Reading inverse operator decomposition from /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis-meg-oct-6-meg-inv.fif...
Reading inverse operator info...
[done]
Reading inverse operator decomposition...
[done]
305 x 305 full covariance (kind = 1) found.
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Noise covariance matrix read.
22494 x 22494 diagonal covariance (kind = 2) found.
Source covariance matrix read.
22494 x 22494 diagonal covariance (kind = 6) found.
Orientation priors read.
22494 x 22494 diagonal covariance (kind = 5) found.
Depth priors read.
Did not find the desired covariance matrix (kind = 3)
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
2 source spaces read
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Source spaces transformed to the inverse solution coordinate frame
Getting helmet for system 306m
Prepare MEG mapping...
Computing dot products for 305 coils...
Computing dot products for 304 surface locations...
Field mapping data ready
Preparing the mapping matrix...
Truncating at 210/305 components to omit less than 0.0001 (9.9e-05)
Channel types:: grad: 203, mag: 102
import os.path as op
import mne
sample_path = mne.datasets.sample.data_path()
subjects_dir = op.join(sample_path, 'subjects')
fname_evoked = op.join(sample_path, 'MEG', 'sample', 'sample_audvis-ave.fif')
fname_inv = op.join(sample_path, 'MEG', 'sample',
'sample_audvis-meg-oct-6-meg-inv.fif')
fname_trans = op.join(sample_path, 'MEG', 'sample',
'sample_audvis_raw-trans.fif')
inv = mne.minimum_norm.read_inverse_operator(fname_inv)
evoked = mne.read_evokeds(fname_evoked, baseline=(None, 0),
proj=True, verbose=False, condition='Left Auditory')
maps = mne.make_field_map(evoked, trans=fname_trans, ch_type='meg',
subject='sample', subjects_dir=subjects_dir)
time = 0.083
fig = mne.viz.create_3d_figure((256, 256))
mne.viz.plot_alignment(
evoked.info, subject='sample', subjects_dir=subjects_dir, fig=fig,
trans=fname_trans, meg='sensors', eeg=False, surfaces='pial',
coord_frame='mri')
evoked.plot_field(maps, time=time, fig=fig, time_label=None, vmax=5e-13)
mne.viz.set_3d_view(
fig, azimuth=40, elevation=87, focalpoint=(0., -0.01, 0.04), roll=-25,
distance=0.55)
Total running time of the script: ( 0 minutes 5.586 seconds)
Estimated memory usage: 9 MB