Note
Click here to download the full example code
Non-parametric 1 sample cluster statistic on single trial power¶
This script shows how to estimate significant clusters in time-frequency power estimates. It uses a non-parametric statistical procedure based on permutations and cluster level statistics.
The procedure consists of:
extracting epochs
compute single trial power estimates
baseline line correct the power estimates (power ratios)
compute stats to see if ratio deviates from 1.
# Authors: Alexandre Gramfort <alexandre.gramfort@inria.fr>
#
# License: BSD-3-Clause
import numpy as np
import matplotlib.pyplot as plt
import mne
from mne.time_frequency import tfr_morlet
from mne.stats import permutation_cluster_1samp_test
from mne.datasets import sample
print(__doc__)
Set parameters¶
data_path = sample.data_path()
raw_fname = data_path + '/MEG/sample/sample_audvis_raw.fif'
tmin, tmax, event_id = -0.3, 0.6, 1
# Setup for reading the raw data
raw = mne.io.read_raw_fif(raw_fname)
events = mne.find_events(raw, stim_channel='STI 014')
include = []
raw.info['bads'] += ['MEG 2443', 'EEG 053'] # bads + 2 more
# picks MEG gradiometers
picks = mne.pick_types(raw.info, meg='grad', eeg=False, eog=True,
stim=False, include=include, exclude='bads')
# Load condition 1
event_id = 1
epochs = mne.Epochs(raw, events, event_id, tmin, tmax, picks=picks,
baseline=(None, 0), preload=True,
reject=dict(grad=4000e-13, eog=150e-6))
# just use right temporal sensors for speed
epochs.pick_channels(mne.read_vectorview_selection('Right-temporal'))
evoked = epochs.average()
# Factor to down-sample the temporal dimension of the TFR computed by
# tfr_morlet. Decimation occurs after frequency decomposition and can
# be used to reduce memory usage (and possibly computational time of downstream
# operations such as nonparametric statistics) if you don't need high
# spectrotemporal resolution.
decim = 5
freqs = np.arange(8, 40, 2) # define frequencies of interest
sfreq = raw.info['sfreq'] # sampling in Hz
tfr_epochs = tfr_morlet(epochs, freqs, n_cycles=4., decim=decim,
average=False, return_itc=False, n_jobs=1)
# Baseline power
tfr_epochs.apply_baseline(mode='logratio', baseline=(-.100, 0))
# Crop in time to keep only what is between 0 and 400 ms
evoked.crop(-0.1, 0.4)
tfr_epochs.crop(-0.1, 0.4)
epochs_power = tfr_epochs.data
Out:
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_raw.fif...
Read a total of 3 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Range : 25800 ... 192599 = 42.956 ... 320.670 secs
Ready.
320 events found
Event IDs: [ 1 2 3 4 5 32]
Not setting metadata
Not setting metadata
72 matching events found
Setting baseline interval to [-0.2996928197375818, 0.0] sec
Applying baseline correction (mode: mean)
3 projection items activated
Loading data for 72 events and 541 original time points ...
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
Rejecting epoch based on EOG : ['EOG 061']
18 bad epochs dropped
Removing projector <Projection | PCA-v1, active : True, n_channels : 102>
Removing projector <Projection | PCA-v2, active : True, n_channels : 102>
Removing projector <Projection | PCA-v3, active : True, n_channels : 102>
Not setting metadata
Applying baseline correction (mode: logratio)
Define adjacency for statistics¶
To compute a cluster-corrected value, we need a suitable definition for the adjacency/adjacency of our values. So we first compute the sensor adjacency, then combine that with a grid/lattice adjacency assumption for the time-frequency plane:
sensor_adjacency, ch_names = mne.channels.find_ch_adjacency(
tfr_epochs.info, 'grad')
# Subselect the channels we are actually using
use_idx = [ch_names.index(ch_name.replace(' ', ''))
for ch_name in tfr_epochs.ch_names]
sensor_adjacency = sensor_adjacency[use_idx][:, use_idx]
assert sensor_adjacency.shape == \
(len(tfr_epochs.ch_names), len(tfr_epochs.ch_names))
assert epochs_power.data.shape == (
len(epochs), len(tfr_epochs.ch_names),
len(tfr_epochs.freqs), len(tfr_epochs.times))
adjacency = mne.stats.combine_adjacency(
sensor_adjacency, len(tfr_epochs.freqs), len(tfr_epochs.times))
# our adjacency is square with each dim matching the data size
assert adjacency.shape[0] == adjacency.shape[1] == \
len(tfr_epochs.ch_names) * len(tfr_epochs.freqs) * len(tfr_epochs.times)
Out:
Reading adjacency matrix for neuromag306planar.
Compute statistic¶
threshold = 3.
n_permutations = 50 # Warning: 50 is way too small for real-world analysis.
T_obs, clusters, cluster_p_values, H0 = \
permutation_cluster_1samp_test(epochs_power, n_permutations=n_permutations,
threshold=threshold, tail=0,
adjacency=adjacency,
out_type='mask', verbose=True)
Out:
stat_fun(H1): min=-6.455144 max=8.265125
Running initial clustering
Found 50 clusters
Permuting 49 times...
0%| | : 0/49 [00:00<?, ?it/s]
2%|2 | : 1/49 [00:00<00:05, 8.67it/s]
4%|4 | : 2/49 [00:00<00:06, 7.52it/s]
6%|6 | : 3/49 [00:00<00:05, 7.98it/s]
8%|8 | : 4/49 [00:00<00:10, 4.36it/s]
10%|# | : 5/49 [00:01<00:08, 4.91it/s]
12%|#2 | : 6/49 [00:01<00:08, 5.35it/s]
14%|#4 | : 7/49 [00:01<00:07, 5.74it/s]
16%|#6 | : 8/49 [00:01<00:06, 6.06it/s]
18%|#8 | : 9/49 [00:01<00:06, 6.34it/s]
20%|## | : 10/49 [00:01<00:07, 5.17it/s]
22%|##2 | : 11/49 [00:02<00:06, 5.44it/s]
24%|##4 | : 12/49 [00:02<00:06, 5.68it/s]
27%|##6 | : 13/49 [00:02<00:06, 5.90it/s]
29%|##8 | : 14/49 [00:02<00:06, 5.14it/s]
31%|### | : 15/49 [00:02<00:06, 5.35it/s]
33%|###2 | : 16/49 [00:02<00:06, 5.33it/s]
35%|###4 | : 17/49 [00:03<00:05, 5.54it/s]
37%|###6 | : 18/49 [00:03<00:05, 5.72it/s]
39%|###8 | : 19/49 [00:03<00:05, 5.08it/s]
41%|#### | : 20/49 [00:03<00:05, 5.16it/s]
43%|####2 | : 21/49 [00:03<00:05, 5.24it/s]
45%|####4 | : 22/49 [00:04<00:05, 5.40it/s]
47%|####6 | : 23/49 [00:04<00:05, 4.99it/s]
49%|####8 | : 24/49 [00:04<00:04, 5.13it/s]
51%|#####1 | : 25/49 [00:04<00:04, 5.28it/s]
53%|#####3 | : 26/49 [00:04<00:04, 5.42it/s]
55%|#####5 | : 27/49 [00:04<00:03, 5.56it/s]
57%|#####7 | : 28/49 [00:05<00:04, 5.08it/s]
59%|#####9 | : 29/49 [00:05<00:03, 5.23it/s]
61%|######1 | : 30/49 [00:05<00:03, 5.30it/s]
63%|######3 | : 31/49 [00:05<00:03, 5.37it/s]
65%|######5 | : 32/49 [00:06<00:03, 5.01it/s]
67%|######7 | : 33/49 [00:06<00:03, 5.15it/s]
69%|######9 | : 34/49 [00:06<00:02, 5.35it/s]
71%|#######1 | : 35/49 [00:06<00:02, 5.48it/s]
73%|#######3 | : 36/49 [00:06<00:02, 5.59it/s]
76%|#######5 | : 37/49 [00:07<00:02, 5.17it/s]
78%|#######7 | : 38/49 [00:07<00:02, 5.30it/s]
80%|#######9 | : 39/49 [00:07<00:01, 5.42it/s]
82%|########1 | : 40/49 [00:07<00:01, 5.54it/s]
84%|########3 | : 41/49 [00:07<00:01, 5.73it/s]
86%|########5 | : 42/49 [00:07<00:01, 5.84it/s]
88%|########7 | : 43/49 [00:07<00:01, 5.40it/s]
90%|########9 | : 44/49 [00:08<00:00, 5.52it/s]
92%|#########1| : 45/49 [00:08<00:00, 5.64it/s]
94%|#########3| : 46/49 [00:08<00:00, 5.76it/s]
96%|#########5| : 47/49 [00:08<00:00, 5.88it/s]
98%|#########7| : 48/49 [00:08<00:00, 5.99it/s]
100%|##########| : 49/49 [00:08<00:00, 6.10it/s]
100%|##########| : 49/49 [00:08<00:00, 5.67it/s]
Computing cluster p-values
Done.
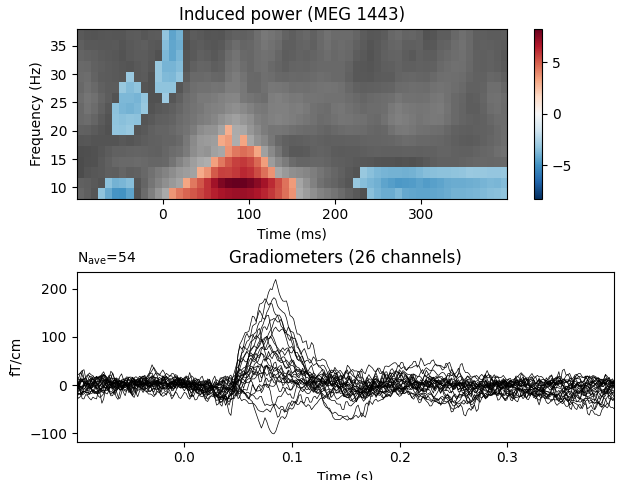
View time-frequency plots¶
evoked_data = evoked.data
times = 1e3 * evoked.times
plt.figure()
plt.subplots_adjust(0.12, 0.08, 0.96, 0.94, 0.2, 0.43)
# Create new stats image with only significant clusters
T_obs_plot = np.nan * np.ones_like(T_obs)
for c, p_val in zip(clusters, cluster_p_values):
if p_val <= 0.05:
T_obs_plot[c] = T_obs[c]
# Just plot one channel's data
ch_idx, f_idx, t_idx = np.unravel_index(
np.nanargmax(np.abs(T_obs_plot)), epochs_power.shape[1:])
# ch_idx = tfr_epochs.ch_names.index('MEG 1332') # to show a specific one
vmax = np.max(np.abs(T_obs))
vmin = -vmax
plt.subplot(2, 1, 1)
plt.imshow(T_obs[ch_idx], cmap=plt.cm.gray,
extent=[times[0], times[-1], freqs[0], freqs[-1]],
aspect='auto', origin='lower', vmin=vmin, vmax=vmax)
plt.imshow(T_obs_plot[ch_idx], cmap=plt.cm.RdBu_r,
extent=[times[0], times[-1], freqs[0], freqs[-1]],
aspect='auto', origin='lower', vmin=vmin, vmax=vmax)
plt.colorbar()
plt.xlabel('Time (ms)')
plt.ylabel('Frequency (Hz)')
plt.title(f'Induced power ({tfr_epochs.ch_names[ch_idx]})')
ax2 = plt.subplot(2, 1, 2)
evoked.plot(axes=[ax2], time_unit='s')
plt.show()
Total running time of the script: ( 0 minutes 15.502 seconds)
Estimated memory usage: 43 MB