Note
Click here to download the full example code
The Epochs data structure: discontinuous data¶
This tutorial covers the basics of creating and working with epoched data. It introduces the Epochs data structure in
detail, including how to load, query, subselect, export, and plot data from an
Epochs object. For more information about visualizing
Epochs objects, see Visualizing epoched data. For info on
creating an Epochs object from (possibly simulated) data in a
NumPy array, see Creating MNE-Python data structures from scratch.
As usual we’ll start by importing the modules we need:
import os
import mne
Epochs objects are a data structure for representing and
analyzing equal-duration chunks of the EEG/MEG signal. Epochs
are most often used to represent data that is time-locked to repeated
experimental events (such as stimulus onsets or subject button presses), but
can also be used for storing sequential or overlapping frames of a continuous
signal (e.g., for analysis of resting-state activity; see
Making equally-spaced Events arrays). Inside an Epochs object, the data
are stored in an array of shape (n_epochs,
n_channels, n_times).
Epochs objects have many similarities with Raw
objects, including:
They can be loaded from and saved to disk in
.fifformat, and their data can be exported to aNumPy arraythrough theget_data()method or to aPandas DataFramethrough theto_data_frame()method.Both
EpochsandRawobjects support channel selection by index or name, includingpick(),pick_channels()andpick_types()methods.SSP projector manipulation is possible through
add_proj(),del_proj(), andplot_projs_topomap()methods.Both
EpochsandRawobjects havecopy(),crop(),time_as_index(),filter(), andresample()methods.Both
EpochsandRawobjects havetimes,ch_names,proj, andinfoattributes.Both
EpochsandRawobjects have built-in plotting methodsplot(),plot_psd(), andplot_psd_topomap().
Creating Epoched data from a Raw object¶
The example dataset we’ve been using thus far doesn’t include pre-epoched
data, so in this section we’ll load the continuous data and create epochs
based on the events recorded in the Raw object’s STIM
channels. As we often do in these tutorials, we’ll crop()
the Raw data to save memory:
sample_data_folder = mne.datasets.sample.data_path()
sample_data_raw_file = os.path.join(sample_data_folder, 'MEG', 'sample',
'sample_audvis_raw.fif')
raw = mne.io.read_raw_fif(sample_data_raw_file, verbose=False).crop(tmax=60)
As we saw in the Parsing events from raw data tutorial, we can extract an
events array from Raw objects using mne.find_events():
events = mne.find_events(raw, stim_channel='STI 014')
Out:
86 events found
Event IDs: [ 1 2 3 4 5 32]
Note
We could also have loaded the events from file, using
mne.read_events():
sample_data_events_file = os.path.join(sample_data_folder,
'MEG', 'sample',
'sample_audvis_raw-eve.fif')
events_from_file = mne.read_events(sample_data_events_file)
See Reading and writing events from/to a file for more details.
The Raw object and the events array are the bare minimum
needed to create an Epochs object, which we create with the
mne.Epochs class constructor. However, you will almost surely want
to change some of the other default parameters. Here we’ll change tmin
and tmax (the time relative to each event at which to start and end each
epoch). Note also that the Epochs constructor accepts
parameters reject and flat for rejecting individual epochs based on
signal amplitude. See the Rejecting Epochs based on channel amplitude section for
examples.
epochs = mne.Epochs(raw, events, tmin=-0.3, tmax=0.7)
Out:
Not setting metadata
Not setting metadata
86 matching events found
Setting baseline interval to [-0.2996928197375818, 0.0] sec
Applying baseline correction (mode: mean)
Created an SSP operator (subspace dimension = 3)
3 projection items activated
You’ll see from the output that:
all 320 events were used to create epochs
baseline correction was automatically applied (by default, baseline is defined as the time span from
tminto0, but can be customized with thebaselineparameter)no additional metadata was provided (see Working with Epoch metadata for details)
the projection operators present in the
Rawfile were copied over to theEpochsobject
If we print the Epochs object, we’ll also see a note that the
epochs are not copied into memory by default, and a count of the number of
epochs created for each integer Event ID.
print(epochs)
Out:
<Epochs | 86 events (good & bad), -0.299693 - 0.699283 sec, baseline -0.299693 – 0 sec, ~3.3 MB, data not loaded,
'1': 20
'2': 20
'3': 20
'32': 4
'4': 18
'5': 4>
Notice that the Event IDs are in quotes; since we didn’t provide an event
dictionary, the mne.Epochs constructor created one automatically and
used the string representation of the integer Event IDs as the dictionary
keys. This is more clear when viewing the event_id attribute:
print(epochs.event_id)
Out:
{'1': 1, '2': 2, '3': 3, '4': 4, '5': 5, '32': 32}
This time let’s pass preload=True and provide an event dictionary; our
provided dictionary will get stored as the event_id attribute and will
make referencing events and pooling across event types easier:
event_dict = {'auditory/left': 1, 'auditory/right': 2, 'visual/left': 3,
'visual/right': 4, 'face': 5, 'buttonpress': 32}
epochs = mne.Epochs(raw, events, tmin=-0.3, tmax=0.7, event_id=event_dict,
preload=True)
print(epochs.event_id)
del raw # we're done with raw, free up some memory
Out:
Not setting metadata
Not setting metadata
86 matching events found
Setting baseline interval to [-0.2996928197375818, 0.0] sec
Applying baseline correction (mode: mean)
Created an SSP operator (subspace dimension = 3)
3 projection items activated
Loading data for 86 events and 601 original time points ...
1 bad epochs dropped
{'auditory/left': 1, 'auditory/right': 2, 'visual/left': 3, 'visual/right': 4, 'face': 5, 'buttonpress': 32}
Notice that the output now mentions “1 bad epoch dropped”. In the tutorial
section Rejecting Epochs based on channel amplitude we saw how you can specify channel
amplitude criteria for rejecting epochs, but here we haven’t specified any
such criteria. In this case, it turns out that the last event was too close
the end of the (cropped) raw file to accommodate our requested tmax of
0.7 seconds, so the final epoch was dropped because it was too short. Here
are the drop_log entries for the last 4 epochs (empty lists indicate
epochs that were not dropped):
print(epochs.drop_log[-4:])
Out:
((), (), (), ('TOO_SHORT',))
Note
If you forget to provide the event dictionary to the Epochs
constructor, you can add it later by assigning to the event_id
attribute:
Basic visualization of Epochs objects¶
The Epochs object can be visualized (and browsed interactively)
using its plot() method:
epochs.plot(n_epochs=10)
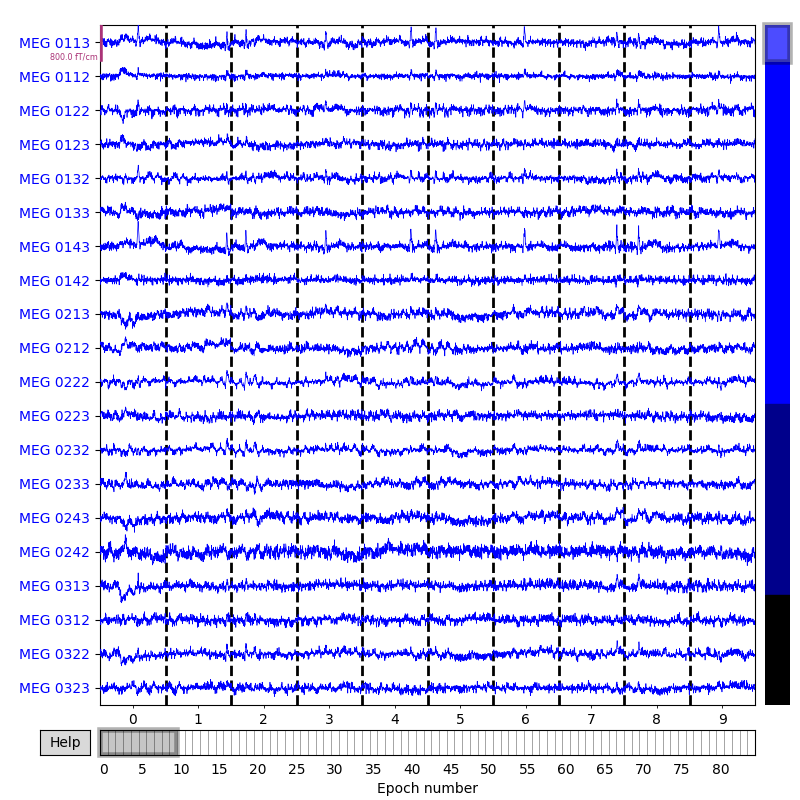
Notice that the individual epochs are sequentially numbered along the bottom
axis and are separated by vertical dashed lines.
Epoch plots are interactive (similar to raw.plot()) and have many of the same interactive controls as
Raw plots. Horizontal and vertical scrollbars allow browsing
through epochs or channels (respectively), and pressing ? when the
plot is focused will show a help screen with all the available controls. See
Visualizing epoched data for more details (as well as other ways of
visualizing epoched data).
Subselecting epochs¶
Now that we have our Epochs object with our descriptive event
labels added, we can subselect epochs easily using square brackets. For
example, we can load all the “catch trials” where the stimulus was a face:
print(epochs['face'])
Out:
<Epochs | 4 events (all good), -0.299693 - 0.699283 sec, baseline -0.299693 – 0 sec, ~10.2 MB, data loaded,
'face': 4>
We can also pool across conditions easily, thanks to how MNE-Python handles
the / character in epoch labels (using what is sometimes called
“tag-based indexing”):
Out:
<Epochs | 40 events (all good), -0.299693 - 0.699283 sec, baseline -0.299693 – 0 sec, ~72.2 MB, data loaded,
'auditory/left': 20
'auditory/right': 20>
<Epochs | 39 events (all good), -0.299693 - 0.699283 sec, baseline -0.299693 – 0 sec, ~70.5 MB, data loaded,
'auditory/left': 20
'visual/left': 19>
You can also pool conditions by passing multiple tags as a list. Note that
MNE-Python will not complain if you ask for tags not present in the object,
as long as it can find some match: the below example is parsed as
(inclusive) 'right' or 'bottom', and you can see from the output
that it selects only auditory/right and visual/right.
print(epochs[['right', 'bottom']])
Out:
<Epochs | 38 events (all good), -0.299693 - 0.699283 sec, baseline -0.299693 – 0 sec, ~68.8 MB, data loaded,
'auditory/right': 20
'visual/right': 18>
However, if no match is found, an error is returned:
try:
print(epochs[['top', 'bottom']])
except KeyError:
print('Tag-based selection with no matches raises a KeyError!')
Out:
Tag-based selection with no matches raises a KeyError!
Selecting epochs by index¶
Epochs objects can also be indexed with integers, slices, or lists of integers. This method of selection ignores event
labels, so if you want the first 10 epochs of a particular type, you can
select the type first, then use integers or slices:
Out:
<Epochs | 10 events (all good), -0.299693 - 0.699283 sec, baseline -0.299693 – 0 sec, ~20.5 MB, data loaded,
'auditory/left': 2
'auditory/right': 3
'visual/left': 3
'visual/right': 2>
<Epochs | 4 events (all good), -0.299693 - 0.699283 sec, baseline -0.299693 – 0 sec, ~10.2 MB, data loaded,
'visual/left': 2
'visual/right': 2>
<Epochs | 4 events (all good), -0.299693 - 0.699283 sec, baseline -0.299693 – 0 sec, ~10.2 MB, data loaded,
'buttonpress': 4>
<Epochs | 4 events (all good), -0.299693 - 0.699283 sec, baseline -0.299693 – 0 sec, ~10.2 MB, data loaded,
'buttonpress': 4>
Selecting, dropping, and reordering channels¶
You can use the pick(), pick_channels(),
pick_types(), and drop_channels() methods
to modify which channels are included in an Epochs object. You
can also use reorder_channels() for this purpose; any
channel names not provided to reorder_channels() will be
dropped. Note that these channel selection methods modify the object
in-place (unlike the square-bracket indexing to select epochs seen above)
so in interactive/exploratory sessions you may want to create a
copy() first.
epochs_eeg = epochs.copy().pick_types(meg=False, eeg=True)
print(epochs_eeg.ch_names)
new_order = ['EEG 002', 'STI 014', 'EOG 061', 'MEG 2521']
epochs_subset = epochs.copy().reorder_channels(new_order)
print(epochs_subset.ch_names)
Out:
Removing projector <Projection | PCA-v1, active : True, n_channels : 102>
Removing projector <Projection | PCA-v2, active : True, n_channels : 102>
Removing projector <Projection | PCA-v3, active : True, n_channels : 102>
['EEG 001', 'EEG 002', 'EEG 003', 'EEG 004', 'EEG 005', 'EEG 006', 'EEG 007', 'EEG 008', 'EEG 009', 'EEG 010', 'EEG 011', 'EEG 012', 'EEG 013', 'EEG 014', 'EEG 015', 'EEG 016', 'EEG 017', 'EEG 018', 'EEG 019', 'EEG 020', 'EEG 021', 'EEG 022', 'EEG 023', 'EEG 024', 'EEG 025', 'EEG 026', 'EEG 027', 'EEG 028', 'EEG 029', 'EEG 030', 'EEG 031', 'EEG 032', 'EEG 033', 'EEG 034', 'EEG 035', 'EEG 036', 'EEG 037', 'EEG 038', 'EEG 039', 'EEG 040', 'EEG 041', 'EEG 042', 'EEG 043', 'EEG 044', 'EEG 045', 'EEG 046', 'EEG 047', 'EEG 048', 'EEG 049', 'EEG 050', 'EEG 051', 'EEG 052', 'EEG 054', 'EEG 055', 'EEG 056', 'EEG 057', 'EEG 058', 'EEG 059', 'EEG 060']
['EEG 002', 'STI 014', 'EOG 061', 'MEG 2521']
del epochs_eeg, epochs_subset
Changing channel name and type¶
You can change the name or type of a channel using
rename_channels() or set_channel_types().
Both methods take dictionaries where the keys are existing
channel names, and the values are the new name (or type) for that channel.
Existing channels that are not in the dictionary will be unchanged.
epochs.rename_channels({'EOG 061': 'BlinkChannel'})
epochs.set_channel_types({'EEG 060': 'ecg'})
print(list(zip(epochs.ch_names, epochs.get_channel_types()))[-4:])
Out:
[('EEG 058', 'eeg'), ('EEG 059', 'eeg'), ('EEG 060', 'ecg'), ('BlinkChannel', 'eog')]
# let's set them back to the correct values before moving on
epochs.rename_channels({'BlinkChannel': 'EOG 061'})
epochs.set_channel_types({'EEG 060': 'eeg'})
Selection in the time domain¶
To change the temporal extent of the Epochs, you can use the
crop() method:
shorter_epochs = epochs.copy().crop(tmin=-0.1, tmax=0.1, include_tmax=True)
for name, obj in dict(Original=epochs, Cropped=shorter_epochs).items():
print('{} epochs has {} time samples'
.format(name, obj.get_data().shape[-1]))
Out:
Original epochs has 601 time samples
Cropped epochs has 121 time samples
Cropping removed part of the baseline. When printing the
cropped Epochs, MNE-Python will inform you about the time
period that was originally used to perform baseline correction by displaying
the string “baseline period cropped after baseline correction”:
print(shorter_epochs)
Out:
<Epochs | 85 events (all good), -0.0998976 - 0.0998976 sec, baseline -0.299693 – 0 sec (baseline period was cropped after baseline correction), ~32.8 MB, data loaded,
'auditory/left': 20
'auditory/right': 20
'buttonpress': 4
'face': 4
'visual/left': 19
'visual/right': 18>
However, if you wanted to expand the time domain of an Epochs
object, you would need to go back to the Raw data and
recreate the Epochs with different values for tmin and/or
tmax.
It is also possible to change the “zero point” that defines the time values
in an Epochs object, with the shift_time()
method. shift_time() allows shifting times relative to the
current values, or specifying a fixed time to set as the new time value of
the first sample (deriving the new time values of subsequent samples based on
the Epochs object’s sampling frequency).
# shift times so that first sample of each epoch is at time zero
later_epochs = epochs.copy().shift_time(tshift=0., relative=False)
print(later_epochs.times[:3])
# shift times by a relative amount
later_epochs.shift_time(tshift=-7, relative=True)
print(later_epochs.times[:3])
Out:
[0. 0.00166496 0.00332992]
[-7. -6.99833504 -6.99667008]
del shorter_epochs, later_epochs
Note that although time shifting respects the sampling frequency (the spacing between samples), it does not enforce the assumption that there is a sample occurring at exactly time=0.
Extracting data in other forms¶
The get_data() method returns the epoched data as a
NumPy array, of shape (n_epochs, n_channels,
n_times); an optional picks parameter selects a subset of channels by
index, name, or type:
eog_data = epochs.get_data(picks='EOG 061')
meg_data = epochs.get_data(picks=['mag', 'grad'])
channel_4_6_8 = epochs.get_data(picks=slice(4, 9, 2))
for name, arr in dict(EOG=eog_data, MEG=meg_data, Slice=channel_4_6_8).items():
print('{} contains {} channels'.format(name, arr.shape[1]))
Out:
EOG contains 1 channels
MEG contains 305 channels
Slice contains 3 channels
Note that if your analysis requires repeatedly extracting single epochs from
an Epochs object, epochs.get_data(item=2) will be much
faster than epochs[2].get_data(), because it avoids the step of
subsetting the Epochs object first.
You can also export Epochs data to Pandas DataFrames. Here, the DataFrame index will be
constructed by converting the time of each sample into milliseconds and
rounding it to the nearest integer, and combining it with the event types and
epoch numbers to form a hierarchical MultiIndex. Each
channel will appear in a separate column. Then you can use any of Pandas’
tools for grouping and aggregating data; for example, here we select any
epochs numbered 10 or less from the auditory/left condition, and extract
times between 100 and 107 ms on channels EEG 056 through EEG 058
(note that slice indexing within Pandas’ loc is
inclusive of the endpoint):
df = epochs.to_data_frame(index=['condition', 'epoch', 'time'])
df.sort_index(inplace=True)
print(df.loc[('auditory/left', slice(0, 10), slice(100, 107)),
'EEG 056':'EEG 058'])
del df
Out:
channel EEG 056 ... EEG 058
condition epoch time ...
auditory/left 2 100 18.814844 ... 16.245426
102 19.172632 ... 16.416629
103 20.186365 ... 17.215579
105 16.787378 ... 14.190982
107 12.970971 ... 11.337589
6 100 1.007473 ... 2.413561
102 -0.602573 ... 1.614611
103 -2.749302 ... 0.701525
105 -7.460178 ... -3.350294
107 -9.726169 ... -5.347669
10 100 4.922387 ... 1.976881
102 1.284874 ... -1.618394
103 0.807824 ... -1.390123
105 4.326073 ... 1.862746
107 5.220543 ... 2.262221
[15 rows x 3 columns]
See the Exporting Epochs to Pandas DataFrames tutorial for many more examples of the
to_data_frame() method.
Loading and saving Epochs objects to disk¶
Epochs objects can be loaded and saved in the .fif format
just like Raw objects, using the mne.read_epochs()
function and the save() method. Functions are also
available for loading data that was epoched outside of MNE-Python, such as
mne.read_epochs_eeglab() and mne.read_epochs_kit().
epochs.save('saved-audiovisual-epo.fif', overwrite=True)
epochs_from_file = mne.read_epochs('saved-audiovisual-epo.fif', preload=False)
Out:
Reading /home/circleci/project/tutorials/epochs/saved-audiovisual-epo.fif ...
Read a total of 3 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Found the data of interest:
t = -299.69 ... 699.28 ms
0 CTF compensation matrices available
Not setting metadata
Not setting metadata
85 matching events found
No baseline correction applied
Created an SSP operator (subspace dimension = 3)
3 projection items activated
The MNE-Python naming convention for epochs files is that the file basename
(the part before the .fif or .fif.gz extension) should end with
-epo or _epo, and a warning will be issued if the filename you
provide does not adhere to that convention.
As a final note, be aware that the class of the epochs object is different
when epochs are loaded from disk rather than generated from a
Raw object:
print(type(epochs))
print(type(epochs_from_file))
Out:
<class 'mne.epochs.Epochs'>
<class 'mne.epochs.EpochsFIF'>
In almost all cases this will not require changing anything about your code. However, if you need to do type checking on epochs objects, you can test against the base class that these classes are derived from:
print(all([isinstance(epochs, mne.BaseEpochs),
isinstance(epochs_from_file, mne.BaseEpochs)]))
Out:
True
Iterating over Epochs¶
Iterating over an Epochs object will yield arrays rather than single-trial Epochs objects:
Out:
<class 'numpy.ndarray'>
<class 'numpy.ndarray'>
<class 'numpy.ndarray'>
If you want to iterate over Epochs objects, you can use an
integer index as the iterator:
Out:
<class 'mne.epochs.Epochs'>
<class 'mne.epochs.Epochs'>
<class 'mne.epochs.Epochs'>
Total running time of the script: ( 0 minutes 20.987 seconds)
Estimated memory usage: 561 MB