Note
Go to the end to download the full example code.
Using contralateral referencing for EEG#
Instead of using a single reference electrode for all channels, some researchers reference the EEG electrodes in each hemisphere to an electrode in the contralateral hemisphere (often an electrode over the mastoid bone; this is common in sleep research for example). Here we demonstrate how to set a contralateral EEG reference.
# Authors: The MNE-Python contributors.
# License: BSD-3-Clause
# Copyright the MNE-Python contributors.
import mne
ssvep_folder = mne.datasets.ssvep.data_path()
ssvep_data_raw_path = (
ssvep_folder / "sub-02" / "ses-01" / "eeg" / "sub-02_ses-01_task-ssvep_eeg.vhdr"
)
raw = mne.io.read_raw(ssvep_data_raw_path, preload=True)
_ = raw.set_montage("easycap-M1")
Extracting parameters from /home/circleci/mne_data/ssvep-example-data/sub-02/ses-01/eeg/sub-02_ses-01_task-ssvep_eeg.vhdr...
Setting channel info structure...
Reading 0 ... 467579 = 0.000 ... 467.579 secs...
The electrodes TP9 and TP10 are near the mastoids so we’ll use them as our contralateral reference channels. Then we’ll create our hemisphere groups.
raw.rename_channels({"TP9": "M1", "TP10": "M2"})
# this splits electrodes into 3 groups; left, midline, and right
ch_names = mne.channels.make_1020_channel_selections(raw.info, return_ch_names=True)
# remove the ref channels from the lists of to-be-rereferenced channels
ch_names["Left"].remove("M1")
ch_names["Right"].remove("M2")
Finally we do the referencing. For the midline channels we’ll reference them to the mean of the two mastoid channels; the left and right hemispheres we’ll reference to the single contralateral mastoid channel.
# midline referencing to mean of mastoids:
mastoids = ["M1", "M2"]
rereferenced_midline_chs = (
raw.copy()
.pick(mastoids + ch_names["Midline"])
.set_eeg_reference(mastoids)
.drop_channels(mastoids)
)
# contralateral referencing (alters channels in `raw` in-place):
for ref, hemi in dict(M2=ch_names["Left"], M1=ch_names["Right"]).items():
mne.set_bipolar_reference(raw, anode=hemi, cathode=[ref] * len(hemi), copy=False)
# strip off '-M1' and '-M2' suffixes added to each bipolar-referenced channel
raw.rename_channels(lambda ch_name: ch_name.split("-")[0])
# replace unreferenced midline with rereferenced midline
_ = raw.drop_channels(ch_names["Midline"]).add_channels([rereferenced_midline_chs])
EEG channel type selected for re-referencing
Applying a custom ('EEG',) reference.
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=13, n_times=467580
Range : 0 ... 467579 = 0.000 ... 467.579 secs
Ready.
Added the following bipolar channels:
O1-M2, PO9-M2, P7-M2, P3-M2, CP5-M2, CP1-M2, T7-M2, C3-M2, FC1-M2, FC5-M2, F3-M2, F7-M2, Fp1-M2
EEG channel type selected for re-referencing
Creating RawArray with float64 data, n_channels=13, n_times=467580
Range : 0 ... 467579 = 0.000 ... 467.579 secs
Ready.
Added the following bipolar channels:
O2-M1, PO10-M1, P8-M1, P4-M1, CP6-M1, CP2-M1, C4-M1, T8-M1, FC2-M1, FC6-M1, F4-M1, F8-M1, Fp2-M1
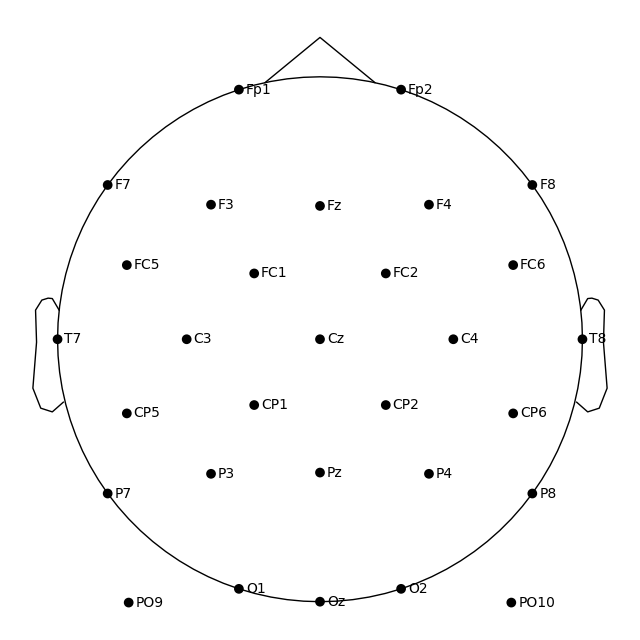
Make sure the channel locations still look right:
fig = raw.plot_sensors(show_names=True, sphere="eeglab")
Approximating Fpz location by mirroring Oz along the X and Y axes.