Note
Go to the end to download the full example code.
Getting impedances from raw files#
Many EEG systems provide impedance measurements for each channel within their file
format. MNE does not parse this information and does not store it in the
Raw object. However, it is possible to extract this information from
the raw data and store it in a separate data structure.
ANT Neuro#
The .cnt file format from ANT Neuro stores impedance information in the form of
triggers. The function mne.io.read_raw_ant() reads this information and marks the
time-segment during which an impedance measurement was performed as
Annotations with the description set in the argument
impedance_annotation. However, it doesn’t extract the impedance values themselves.
To do so, use the function antio.parser.read_triggers.
# Authors: The MNE-Python contributors.
# License: BSD-3-Clause
# Copyright the MNE-Python contributors.
from antio import read_cnt
from antio.parser import read_triggers
from matplotlib import pyplot as plt
from mne.datasets import testing
from mne.io import read_raw_ant
from mne.viz import plot_topomap
fname = testing.data_path() / "antio" / "CA_208" / "test_CA_208.cnt"
cnt = read_cnt(fname)
_, _, _, impedances, _ = read_triggers(cnt)
raw = read_raw_ant(fname, eog=r"EOG")
impedances = [{ch: imp[k] for k, ch in enumerate(raw.ch_names)} for imp in impedances]
print(impedances[0]) # impedances measurement at the beginning of the recording
Reading ANT file /home/circleci/mne_data/MNE-testing-data/antio/CA_208/test_CA_208.cnt
All 63 EEG channels are referenced to CPz.
{'Fp1': 2.254, 'Fpz': 2.926, 'Fp2': 2.693, 'F7': 13.045, 'F3': 0.902, 'Fz': 2.737, 'F4': 0.149, 'F8': 25.132999, 'FC5': 0.304, 'FC1': 38.924, 'FC2': 2.258, 'FC6': 0.135, 'M1': 4.445, 'T7': 6.233, 'C3': 8.322, 'Cz': 0.064, 'C4': 1.271, 'T8': 4.049, 'M2': 1.846, 'CP5': 2.721, 'CP1': 2.944, 'CP2': 1.218, 'CP6': 0.879, 'P7': 22.726999, 'P3': 9.176, 'Pz': 1.84, 'P4': 27.167, 'P8': 1.157, 'POz': 3.575, 'O1': 2.698, 'O2': 8.073, 'EOG': 15.205, 'AF7': 5.842, 'AF3': 1.303, 'AF4': 2.136, 'AF8': 0.787, 'F5': 0.863, 'F1': 1.696, 'F2': 1.616, 'F6': 0.478, 'FC3': 6.01, 'FCz': 0.72, 'FC4': 4.314, 'C5': 10.945, 'C1': 6.151, 'C2': 0.772, 'C6': 0.879, 'CP3': 12.115, 'CP4': 1.887, 'P5': 14.045, 'P1': 2.733, 'P2': 34.641998, 'P6': 1.765, 'PO5': 5.666, 'PO3': 14.12, 'PO4': 2.599, 'PO6': 11.339, 'FT7': 13.158, 'FT8': 1.046, 'TP7': 5.917, 'TP8': 2.61, 'PO7': 5.691, 'PO8': 1.759, 'Oz': 5.165, 'BIP1': 2147483.75, 'BIP2': 2147483.75, 'BIP3': 2147483.75, 'BIP4': 2147483.75, 'BIP5': 2147483.75, 'BIP6': 2147483.75, 'BIP7': 2147483.75, 'BIP8': 2147483.75, 'BIP9': 2147483.75, 'BIP10': 2147483.75, 'BIP11': 2147483.75, 'BIP12': 2147483.75, 'BIP13': 2147483.75, 'BIP14': 2147483.75, 'BIP15': 2147483.75, 'BIP16': 2147483.75, 'BIP17': 2147483.75, 'BIP18': 2147483.75, 'BIP19': 2147483.75, 'BIP20': 2147483.75, 'BIP21': 2147483.75, 'BIP22': 2147483.75, 'BIP23': 2147483.75, 'BIP24': 2147483.75}
Note that the impedance measurement contains all channels, including the bipolar ones. We can visualize the impedances on a topographic map; below we show a topography of impedances before and after the recording for the EEG channels only.
raw.pick("eeg").set_montage("standard_1020")
impedances = [{ch: imp[ch] for ch in raw.ch_names} for imp in impedances]
f, ax = plt.subplots(1, 2, layout="constrained", figsize=(10, 5))
f.suptitle("Impedances (kOhm)")
impedance = list(impedances[0].values())
plot_topomap(
impedance,
raw.info,
vlim=(0, 50),
axes=ax[0],
show=False,
names=[f"{elt:.1f}" for elt in impedance],
)
ax[0].set_title("Impedances at the beginning of the recording")
impedance = list(impedances[-1].values())
plot_topomap(
impedance,
raw.info,
vlim=(0, 50),
axes=ax[1],
show=False,
names=[f"{elt:.1f}" for elt in impedance],
)
ax[1].set_title("Impedances at the end of the recording")
plt.show()
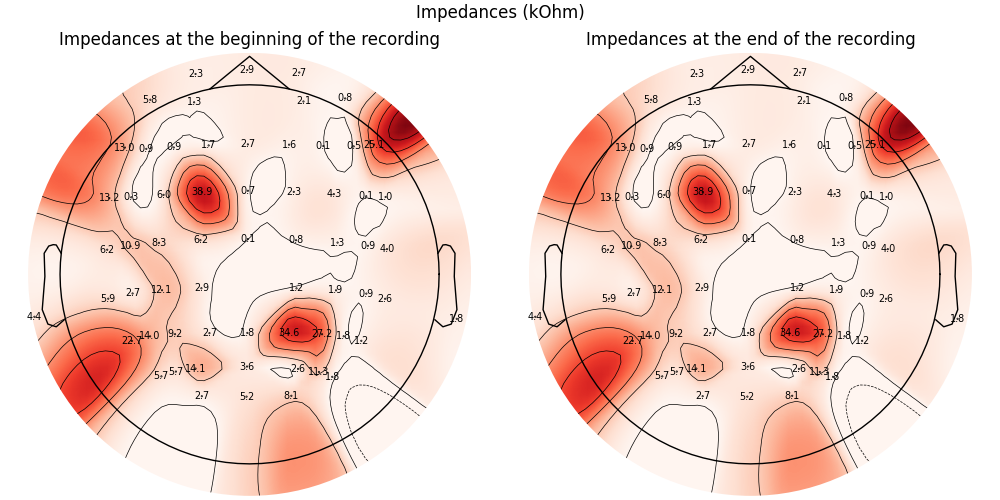
In this very short test file, the impedances are stable over time.
Total running time of the script: (0 minutes 1.682 seconds)