Note
Click here to download the full example code
Motor imagery decoding from EEG data using the Common Spatial Pattern (CSP)#
Decoding of motor imagery applied to EEG data decomposed using CSP. A classifier is then applied to features extracted on CSP-filtered signals.
See https://en.wikipedia.org/wiki/Common_spatial_pattern and 1. The EEGBCI dataset is documented in 2 and is available at PhysioNet 3.
# Authors: Martin Billinger <martin.billinger@tugraz.at>
#
# License: BSD-3-Clause
import numpy as np
import matplotlib.pyplot as plt
from sklearn.pipeline import Pipeline
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.model_selection import ShuffleSplit, cross_val_score
from mne import Epochs, pick_types, events_from_annotations
from mne.channels import make_standard_montage
from mne.io import concatenate_raws, read_raw_edf
from mne.datasets import eegbci
from mne.decoding import CSP
print(__doc__)
# #############################################################################
# # Set parameters and read data
# avoid classification of evoked responses by using epochs that start 1s after
# cue onset.
tmin, tmax = -1., 4.
event_id = dict(hands=2, feet=3)
subject = 1
runs = [6, 10, 14] # motor imagery: hands vs feet
raw_fnames = eegbci.load_data(subject, runs)
raw = concatenate_raws([read_raw_edf(f, preload=True) for f in raw_fnames])
eegbci.standardize(raw) # set channel names
montage = make_standard_montage('standard_1005')
raw.set_montage(montage)
# strip channel names of "." characters
raw.rename_channels(lambda x: x.strip('.'))
# Apply band-pass filter
raw.filter(7., 30., fir_design='firwin', skip_by_annotation='edge')
events, _ = events_from_annotations(raw, event_id=dict(T1=2, T2=3))
picks = pick_types(raw.info, meg=False, eeg=True, stim=False, eog=False,
exclude='bads')
# Read epochs (train will be done only between 1 and 2s)
# Testing will be done with a running classifier
epochs = Epochs(raw, events, event_id, tmin, tmax, proj=True, picks=picks,
baseline=None, preload=True)
epochs_train = epochs.copy().crop(tmin=1., tmax=2.)
labels = epochs.events[:, -1] - 2
Extracting EDF parameters from /home/circleci/mne_data/MNE-eegbci-data/files/eegmmidb/1.0.0/S001/S001R06.edf...
EDF file detected
Setting channel info structure...
Creating raw.info structure...
Reading 0 ... 19999 = 0.000 ... 124.994 secs...
Extracting EDF parameters from /home/circleci/mne_data/MNE-eegbci-data/files/eegmmidb/1.0.0/S001/S001R10.edf...
EDF file detected
Setting channel info structure...
Creating raw.info structure...
Reading 0 ... 19999 = 0.000 ... 124.994 secs...
Extracting EDF parameters from /home/circleci/mne_data/MNE-eegbci-data/files/eegmmidb/1.0.0/S001/S001R14.edf...
EDF file detected
Setting channel info structure...
Creating raw.info structure...
Reading 0 ... 19999 = 0.000 ... 124.994 secs...
Filtering raw data in 3 contiguous segments
Setting up band-pass filter from 7 - 30 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 7.00
- Lower transition bandwidth: 2.00 Hz (-6 dB cutoff frequency: 6.00 Hz)
- Upper passband edge: 30.00 Hz
- Upper transition bandwidth: 7.50 Hz (-6 dB cutoff frequency: 33.75 Hz)
- Filter length: 265 samples (1.656 sec)
Used Annotations descriptions: ['T1', 'T2']
Not setting metadata
45 matching events found
No baseline correction applied
0 projection items activated
Using data from preloaded Raw for 45 events and 801 original time points ...
0 bad epochs dropped
Classification with linear discrimant analysis
# Define a monte-carlo cross-validation generator (reduce variance):
scores = []
epochs_data = epochs.get_data()
epochs_data_train = epochs_train.get_data()
cv = ShuffleSplit(10, test_size=0.2, random_state=42)
cv_split = cv.split(epochs_data_train)
# Assemble a classifier
lda = LinearDiscriminantAnalysis()
csp = CSP(n_components=4, reg=None, log=True, norm_trace=False)
# Use scikit-learn Pipeline with cross_val_score function
clf = Pipeline([('CSP', csp), ('LDA', lda)])
scores = cross_val_score(clf, epochs_data_train, labels, cv=cv, n_jobs=1)
# Printing the results
class_balance = np.mean(labels == labels[0])
class_balance = max(class_balance, 1. - class_balance)
print("Classification accuracy: %f / Chance level: %f" % (np.mean(scores),
class_balance))
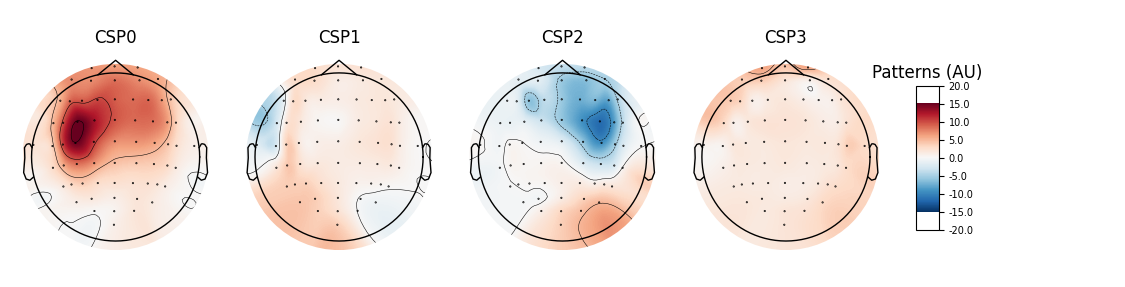
# plot CSP patterns estimated on full data for visualization
csp.fit_transform(epochs_data, labels)
csp.plot_patterns(epochs.info, ch_type='eeg', units='Patterns (AU)', size=1.5)
Computing rank from data with rank=None
Using tolerance 9.7e-05 (2.2e-16 eps * 64 dim * 6.8e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.3e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 9.5e-05 (2.2e-16 eps * 64 dim * 6.7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.2e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00012 (2.2e-16 eps * 64 dim * 8.6e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 9.6e-05 (2.2e-16 eps * 64 dim * 6.7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00011 (2.2e-16 eps * 64 dim * 7.5e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00011 (2.2e-16 eps * 64 dim * 7.6e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00012 (2.2e-16 eps * 64 dim * 8.1e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.2e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00012 (2.2e-16 eps * 64 dim * 8.5e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 9.5e-05 (2.2e-16 eps * 64 dim * 6.7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00012 (2.2e-16 eps * 64 dim * 8.6e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 9.6e-05 (2.2e-16 eps * 64 dim * 6.7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00012 (2.2e-16 eps * 64 dim * 8.3e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.2e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.1e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00011 (2.2e-16 eps * 64 dim * 7.7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.3e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Classification accuracy: 0.933333 / Chance level: 0.533333
Computing rank from data with rank=None
Using tolerance 0.00025 (2.2e-16 eps * 64 dim * 1.7e+10 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00026 (2.2e-16 eps * 64 dim * 1.9e+10 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
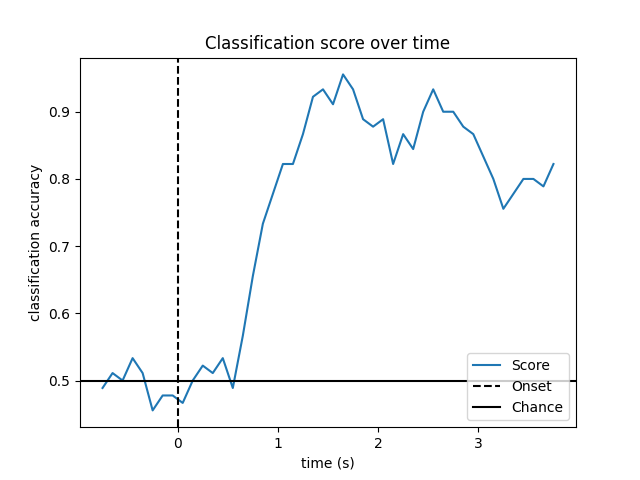
Look at performance over time
sfreq = raw.info['sfreq']
w_length = int(sfreq * 0.5) # running classifier: window length
w_step = int(sfreq * 0.1) # running classifier: window step size
w_start = np.arange(0, epochs_data.shape[2] - w_length, w_step)
scores_windows = []
for train_idx, test_idx in cv_split:
y_train, y_test = labels[train_idx], labels[test_idx]
X_train = csp.fit_transform(epochs_data_train[train_idx], y_train)
X_test = csp.transform(epochs_data_train[test_idx])
# fit classifier
lda.fit(X_train, y_train)
# running classifier: test classifier on sliding window
score_this_window = []
for n in w_start:
X_test = csp.transform(epochs_data[test_idx][:, :, n:(n + w_length)])
score_this_window.append(lda.score(X_test, y_test))
scores_windows.append(score_this_window)
# Plot scores over time
w_times = (w_start + w_length / 2.) / sfreq + epochs.tmin
plt.figure()
plt.plot(w_times, np.mean(scores_windows, 0), label='Score')
plt.axvline(0, linestyle='--', color='k', label='Onset')
plt.axhline(0.5, linestyle='-', color='k', label='Chance')
plt.xlabel('time (s)')
plt.ylabel('classification accuracy')
plt.title('Classification score over time')
plt.legend(loc='lower right')
plt.show()
Computing rank from data with rank=None
Using tolerance 9.7e-05 (2.2e-16 eps * 64 dim * 6.8e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.3e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 9.5e-05 (2.2e-16 eps * 64 dim * 6.7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.2e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00012 (2.2e-16 eps * 64 dim * 8.6e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 9.6e-05 (2.2e-16 eps * 64 dim * 6.7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00011 (2.2e-16 eps * 64 dim * 7.5e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00011 (2.2e-16 eps * 64 dim * 7.6e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00012 (2.2e-16 eps * 64 dim * 8.1e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.2e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00012 (2.2e-16 eps * 64 dim * 8.5e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 9.5e-05 (2.2e-16 eps * 64 dim * 6.7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00012 (2.2e-16 eps * 64 dim * 8.6e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 9.6e-05 (2.2e-16 eps * 64 dim * 6.7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00012 (2.2e-16 eps * 64 dim * 8.3e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.2e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.1e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.00011 (2.2e-16 eps * 64 dim * 7.7e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
Computing rank from data with rank=None
Using tolerance 0.0001 (2.2e-16 eps * 64 dim * 7.3e+09 max singular value)
Estimated rank (mag): 64
MAG: rank 64 computed from 64 data channels with 0 projectors
Reducing data rank from 64 -> 64
Estimating covariance using EMPIRICAL
Done.
References#
- 1
Zoltan J. Koles. The quantitative extraction and topographic mapping of the abnormal components in the clinical EEG. Electroencephalography and Clinical Neurophysiology, 79(6):440–447, 1991. doi:10.1016/0013-4694(91)90163-X.
- 2
Gerwin Schalk, Dennis J. McFarland, Thilo Hinterberger, Niels Birbaumer, and Jonathan R. Wolpaw. BCI2000: a general-purpose brain-computer interface (BCI) system. IEEE Transactions on Biomedical Engineering, 51(6):1034–1043, 2004. doi:10.1109/TBME.2004.827072.
- 3
Ary L. Goldberger, Luis A. N. Amaral, Leon Glass, Jeffrey M. Hausdorff, Plamen Ch. Ivanov, Roger G. Mark, Joseph E. Mietus, George B. Moody, Chung-Kang Peng, and H. Eugene Stanley. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation, 2000. doi:10.1161/01.CIR.101.23.e215.
Total running time of the script: ( 0 minutes 7.160 seconds)
Estimated memory usage: 9 MB