Note
Click here to download the full example code
Source localization with equivalent current dipole (ECD) fit#
This shows how to fit a dipole 1 using mne-python.
For a comparison of fits between MNE-C and MNE-Python, see this gist.
import os.path as op
import numpy as np
import matplotlib.pyplot as plt
import mne
from mne.forward import make_forward_dipole
from mne.evoked import combine_evoked
from mne.simulation import simulate_evoked
from nilearn.plotting import plot_anat
from nilearn.datasets import load_mni152_template
data_path = mne.datasets.sample.data_path()
subjects_dir = op.join(data_path, 'subjects')
fname_ave = op.join(data_path, 'MEG', 'sample', 'sample_audvis-ave.fif')
fname_cov = op.join(data_path, 'MEG', 'sample', 'sample_audvis-cov.fif')
fname_bem = op.join(subjects_dir, 'sample', 'bem', 'sample-5120-bem-sol.fif')
fname_trans = op.join(data_path, 'MEG', 'sample',
'sample_audvis_raw-trans.fif')
fname_surf_lh = op.join(subjects_dir, 'sample', 'surf', 'lh.white')
Let’s localize the N100m (using MEG only)
evoked = mne.read_evokeds(fname_ave, condition='Right Auditory',
baseline=(None, 0))
evoked.pick_types(meg=True, eeg=False)
evoked_full = evoked.copy()
evoked.crop(0.07, 0.08)
# Fit a dipole
dip = mne.fit_dipole(evoked, fname_cov, fname_bem, fname_trans)[0]
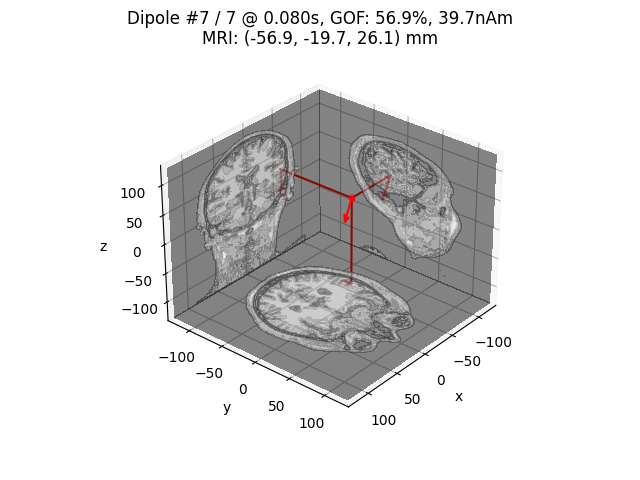
# Plot the result in 3D brain with the MRI image.
dip.plot_locations(fname_trans, 'sample', subjects_dir, mode='orthoview')
Reading /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis-ave.fif ...
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Found the data of interest:
t = -199.80 ... 499.49 ms (Right Auditory)
0 CTF compensation matrices available
nave = 61 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
Applying baseline correction (mode: mean)
Removing projector <Projection | Average EEG reference, active : True, n_channels : 60>
MRI transform : /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_raw-trans.fif
Head origin : -4.3 18.4 67.0 mm rad = 71.8 mm.
Guess grid : 20.0 mm
Guess mindist : 5.0 mm
Guess exclude : 20.0 mm
Using normal MEG coil definitions.
Noise covariance : /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis-cov.fif
Coordinate transformation: MRI (surface RAS) -> head
0.999310 0.009985 -0.035787 -3.17 mm
0.012759 0.812405 0.582954 6.86 mm
0.034894 -0.583008 0.811716 28.88 mm
0.000000 0.000000 0.000000 1.00
Coordinate transformation: MEG device -> head
0.991420 -0.039936 -0.124467 -6.13 mm
0.060661 0.984012 0.167456 0.06 mm
0.115790 -0.173570 0.977991 64.74 mm
0.000000 0.000000 0.000000 1.00
0 bad channels total
Read 305 MEG channels from info
105 coil definitions read
Coordinate transformation: MEG device -> head
0.991420 -0.039936 -0.124467 -6.13 mm
0.060661 0.984012 0.167456 0.06 mm
0.115790 -0.173570 0.977991 64.74 mm
0.000000 0.000000 0.000000 1.00
MEG coil definitions created in head coordinates.
Decomposing the sensor noise covariance matrix...
Created an SSP operator (subspace dimension = 3)
Computing rank from covariance with rank=None
Using tolerance 3.3e-13 (2.2e-16 eps * 305 dim * 4.8 max singular value)
Estimated rank (mag + grad): 302
MEG: rank 302 computed from 305 data channels with 3 projectors
Setting small MEG eigenvalues to zero (without PCA)
Created the whitener using a noise covariance matrix with rank 302 (3 small eigenvalues omitted)
---- Computing the forward solution for the guesses...
Guess surface (inner skull) is in MRI (surface RAS) coordinates
Filtering (grid = 20 mm)...
Surface CM = ( 0.7 -10.0 44.3) mm
Surface fits inside a sphere with radius 91.8 mm
Surface extent:
x = -66.7 ... 68.8 mm
y = -88.0 ... 79.0 mm
z = -44.5 ... 105.8 mm
Grid extent:
x = -80.0 ... 80.0 mm
y = -100.0 ... 80.0 mm
z = -60.0 ... 120.0 mm
900 sources before omitting any.
396 sources after omitting infeasible sources not within 20.0 - 91.8 mm.
Source spaces are in MRI coordinates.
Checking that the sources are inside the surface and at least 5.0 mm away (will take a few...)
Checking surface interior status for 396 points...
Found 42/396 points inside an interior sphere of radius 43.6 mm
Found 0/396 points outside an exterior sphere of radius 91.8 mm
Found 186/354 points outside using surface Qhull
Found 9/168 points outside using solid angles
Total 201/396 points inside the surface
Interior check completed in 129.0 ms
195 source space points omitted because they are outside the inner skull surface.
45 source space points omitted because of the 5.0-mm distance limit.
156 sources remaining after excluding the sources outside the surface and less than 5.0 mm inside.
Go through all guess source locations...
[done 156 sources]
---- Fitted : 69.9 ms, distance to inner skull : 10.7283 mm
---- Fitted : 71.6 ms, distance to inner skull : 10.6265 mm
---- Fitted : 73.3 ms, distance to inner skull : 10.5323 mm
---- Fitted : 74.9 ms, distance to inner skull : 10.3372 mm
---- Fitted : 76.6 ms, distance to inner skull : 9.9420 mm
---- Fitted : 78.3 ms, distance to inner skull : 9.7364 mm
---- Fitted : 79.9 ms, distance to inner skull : 9.4769 mm
Projections have already been applied. Setting proj attribute to True.
7 time points fitted
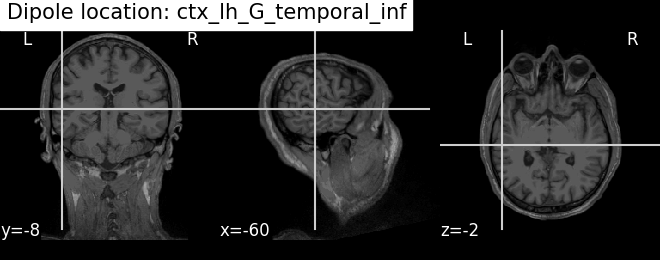
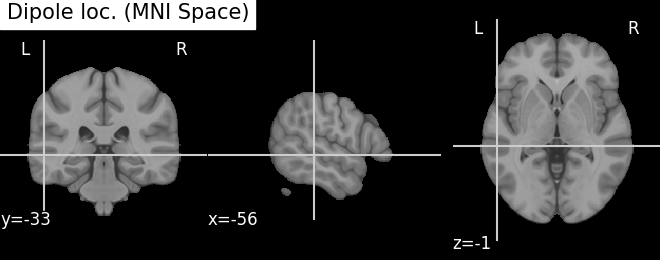
Plot the result in 3D brain with the MRI image using Nilearn In MRI coordinates and in MNI coordinates (template brain)
subject = 'sample'
mni_pos = dip.to_mni(subject=subject, trans=fname_trans,
subjects_dir=subjects_dir)
mri_pos = dip.to_mri(subject=subject, trans=fname_trans,
subjects_dir=subjects_dir)
# Find an anatomical label for the best fitted dipole
best_dip_idx = dip.gof.argmax()
label = dip.to_volume_labels(fname_trans, subject=subject,
subjects_dir=subjects_dir,
aseg='aparc.a2009s+aseg')[best_dip_idx]
# Draw dipole position on MRI scan and add anatomical label from parcellation
t1_fname = op.join(subjects_dir, subject, 'mri', 'T1.mgz')
fig_T1 = plot_anat(t1_fname, cut_coords=mri_pos[0],
title=f'Dipole location: {label}')
try:
template = load_mni152_template(resolution=1)
except TypeError: # in nilearn < 0.8.1 this did not exist
template = load_mni152_template()
fig_template = plot_anat(template, cut_coords=mni_pos[0],
title='Dipole loc. (MNI Space)')
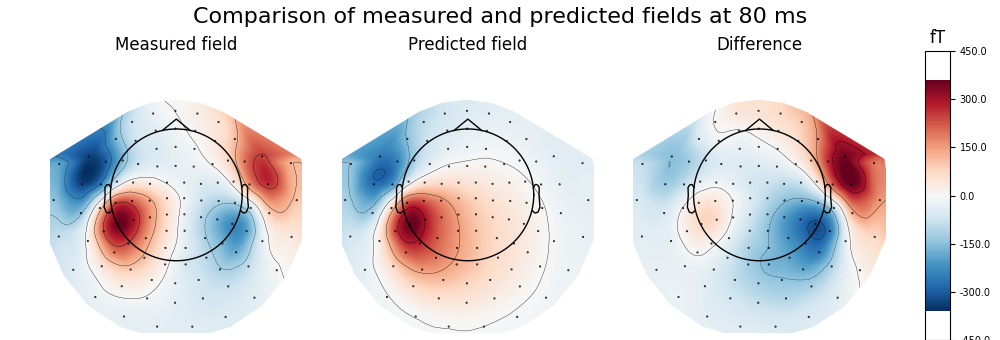
Calculate and visualise magnetic field predicted by dipole with maximum GOF and compare to the measured data, highlighting the ipsilateral (right) source
fwd, stc = make_forward_dipole(dip, fname_bem, evoked.info, fname_trans)
pred_evoked = simulate_evoked(fwd, stc, evoked.info, cov=None, nave=np.inf)
# find time point with highest GOF to plot
best_idx = np.argmax(dip.gof)
best_time = dip.times[best_idx]
print('Highest GOF %0.1f%% at t=%0.1f ms with confidence volume %0.1f cm^3'
% (dip.gof[best_idx], best_time * 1000,
dip.conf['vol'][best_idx] * 100 ** 3))
# remember to create a subplot for the colorbar
fig, axes = plt.subplots(nrows=1, ncols=4, figsize=[10., 3.4],
gridspec_kw=dict(width_ratios=[1, 1, 1, 0.1],
top=0.85))
vmin, vmax = -400, 400 # make sure each plot has same colour range
# first plot the topography at the time of the best fitting (single) dipole
plot_params = dict(times=best_time, ch_type='mag', outlines='skirt',
colorbar=False, time_unit='s')
evoked.plot_topomap(time_format='Measured field', axes=axes[0], **plot_params)
# compare this to the predicted field
pred_evoked.plot_topomap(time_format='Predicted field', axes=axes[1],
**plot_params)
# Subtract predicted from measured data (apply equal weights)
diff = combine_evoked([evoked, pred_evoked], weights=[1, -1])
plot_params['colorbar'] = True
diff.plot_topomap(time_format='Difference', axes=axes[2:], **plot_params)
fig.suptitle('Comparison of measured and predicted fields '
'at {:.0f} ms'.format(best_time * 1000.), fontsize=16)
fig.tight_layout()
Positions (in meters) and orientations
7 sources
Source space : <SourceSpaces: [<discrete, n_used=7>] head coords, ~3 kB>
MRI -> head transform : /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_raw-trans.fif
Measurement data : instance of Info
Conductor model : /home/circleci/mne_data/MNE-sample-data/subjects/sample/bem/sample-5120-bem-sol.fif
Accurate field computations
Do computations in head coordinates
Free source orientations
Read 1 source spaces a total of 7 active source locations
Coordinate transformation: MRI (surface RAS) -> head
0.999310 0.009985 -0.035787 -3.17 mm
0.012759 0.812405 0.582954 6.86 mm
0.034894 -0.583008 0.811716 28.88 mm
0.000000 0.000000 0.000000 1.00
Read 305 MEG channels from info
105 coil definitions read
Coordinate transformation: MEG device -> head
0.991420 -0.039936 -0.124467 -6.13 mm
0.060661 0.984012 0.167456 0.06 mm
0.115790 -0.173570 0.977991 64.74 mm
0.000000 0.000000 0.000000 1.00
MEG coil definitions created in head coordinates.
Source spaces are now in head coordinates.
Setting up the BEM model using /home/circleci/mne_data/MNE-sample-data/subjects/sample/bem/sample-5120-bem-sol.fif...
Loading surfaces...
Loading the solution matrix...
Homogeneous model surface loaded.
Loaded linear_collocation BEM solution from /home/circleci/mne_data/MNE-sample-data/subjects/sample/bem/sample-5120-bem-sol.fif
Employing the head->MRI coordinate transform with the BEM model.
BEM model sample-5120-bem-sol.fif is now set up
Source spaces are in head coordinates.
Checking that the sources are inside the surface (will take a few...)
Checking surface interior status for 7 points...
Found 0/7 points inside an interior sphere of radius 43.6 mm
Found 0/7 points outside an exterior sphere of radius 91.8 mm
Found 0/7 points outside using surface Qhull
Found 0/7 points outside using solid angles
Total 7/7 points inside the surface
Interior check completed in 71.8 ms
Checking surface interior status for 305 points...
Found 0/305 points inside an interior sphere of radius 43.6 mm
Found 305/305 points outside an exterior sphere of radius 91.8 mm
Found 0/ 0 points outside using surface Qhull
Found 0/ 0 points outside using solid angles
Total 0/305 points inside the surface
Interior check completed in 21.7 ms
Setting up compensation data...
No compensation set. Nothing more to do.
Composing the field computation matrix...
Computing MEG at 7 source locations (free orientations)...
Finished.
Changing to fixed-orientation forward solution with surface-based source orientations...
[done]
Projecting source estimate to sensor space...
[done]
Highest GOF 56.9% at t=79.9 ms with confidence volume 8.1 cm^3
/home/circleci/project/tutorials/inverse/20_dipole_fit.py:114: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
fig.tight_layout()
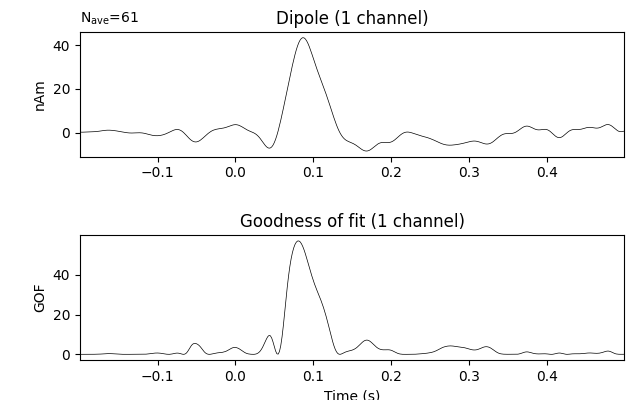
Estimate the time course of a single dipole with fixed position and orientation (the one that maximized GOF) over the entire interval
dip_fixed = mne.fit_dipole(evoked_full, fname_cov, fname_bem, fname_trans,
pos=dip.pos[best_idx], ori=dip.ori[best_idx])[0]
dip_fixed.plot(time_unit='s')
MRI transform : /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_raw-trans.fif
Head origin : -4.3 18.4 67.0 mm rad = 71.8 mm.
Fixed position : -61.1 5.3 59.6 mm
Fixed orientation : 0.0113 -0.7506 -0.6606 mm
Noise covariance : /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis-cov.fif
Coordinate transformation: MRI (surface RAS) -> head
0.999310 0.009985 -0.035787 -3.17 mm
0.012759 0.812405 0.582954 6.86 mm
0.034894 -0.583008 0.811716 28.88 mm
0.000000 0.000000 0.000000 1.00
Coordinate transformation: MEG device -> head
0.991420 -0.039936 -0.124467 -6.13 mm
0.060661 0.984012 0.167456 0.06 mm
0.115790 -0.173570 0.977991 64.74 mm
0.000000 0.000000 0.000000 1.00
0 bad channels total
Read 305 MEG channels from info
105 coil definitions read
Coordinate transformation: MEG device -> head
0.991420 -0.039936 -0.124467 -6.13 mm
0.060661 0.984012 0.167456 0.06 mm
0.115790 -0.173570 0.977991 64.74 mm
0.000000 0.000000 0.000000 1.00
MEG coil definitions created in head coordinates.
Decomposing the sensor noise covariance matrix...
Created an SSP operator (subspace dimension = 3)
Computing rank from covariance with rank=None
Using tolerance 3.3e-13 (2.2e-16 eps * 305 dim * 4.8 max singular value)
Estimated rank (mag + grad): 302
MEG: rank 302 computed from 305 data channels with 3 projectors
Setting small MEG eigenvalues to zero (without PCA)
Created the whitener using a noise covariance matrix with rank 302 (3 small eigenvalues omitted)
Compute forward for dipole location...
[done 1 source]
Projections have already been applied. Setting proj attribute to True.
421 time points fitted
References#
- 1
Jukka Sarvas. Basic mathematical and electromagnetic concepts of the biomagnetic inverse problem. Physics in Medicine and Biology, 32(1):11–22, 1987. doi:10.1088/0031-9155/32/1/004.
Total running time of the script: ( 0 minutes 35.292 seconds)
Estimated memory usage: 304 MB