Note
Click here to download the full example code
Plot a cortical parcellation#
In this example, we download the HCP-MMP1.0 parcellation
1 and show it on fsaverage.
We will also download the customized 448-label aparc
parcellation from 2.
Note
The HCP-MMP dataset has license terms restricting its use. Of particular relevance:
“I will acknowledge the use of WU-Minn HCP data and data derived from WU-Minn HCP data when publicly presenting any results or algorithms that benefitted from their use.”
# Author: Eric Larson <larson.eric.d@gmail.com>
# Denis Engemann <denis.engemann@gmail.com>
#
# License: BSD-3-Clause
import mne
Brain = mne.viz.get_brain_class()
subjects_dir = mne.datasets.sample.data_path() / 'subjects'
mne.datasets.fetch_hcp_mmp_parcellation(subjects_dir=subjects_dir,
verbose=True)
mne.datasets.fetch_aparc_sub_parcellation(subjects_dir=subjects_dir,
verbose=True)
labels = mne.read_labels_from_annot(
'fsaverage', 'HCPMMP1', 'lh', subjects_dir=subjects_dir)
brain = Brain('fsaverage', 'lh', 'inflated', subjects_dir=subjects_dir,
cortex='low_contrast', background='white', size=(800, 600))
brain.add_annotation('HCPMMP1')
aud_label = [label for label in labels if label.name == 'L_A1_ROI-lh'][0]
brain.add_label(aud_label, borders=False)
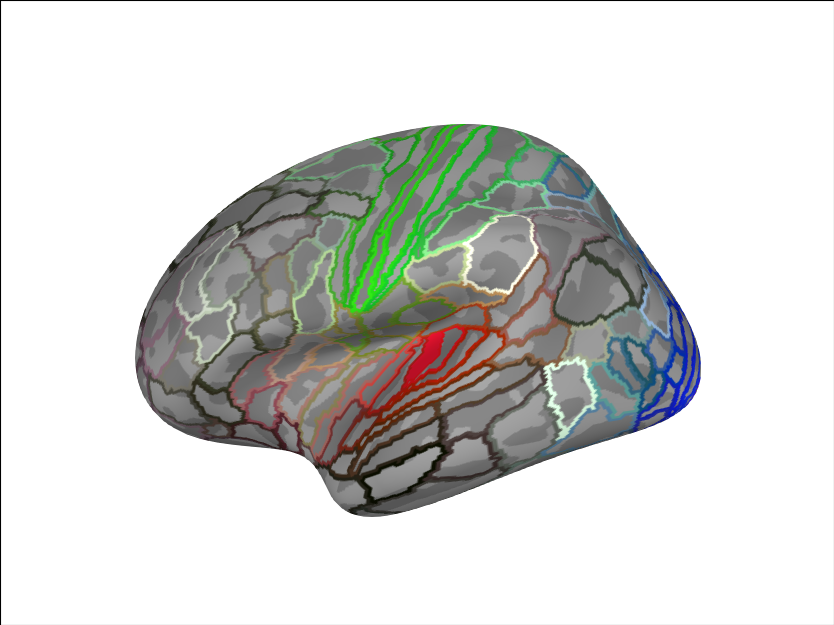
Reading labels from parcellation...
read 181 labels from /home/circleci/mne_data/MNE-sample-data/subjects/fsaverage/label/lh.HCPMMP1.annot
We can also plot a combined set of labels (23 per hemisphere).
brain = Brain('fsaverage', 'lh', 'inflated', subjects_dir=subjects_dir,
cortex='low_contrast', background='white', size=(800, 600))
brain.add_annotation('HCPMMP1_combined')
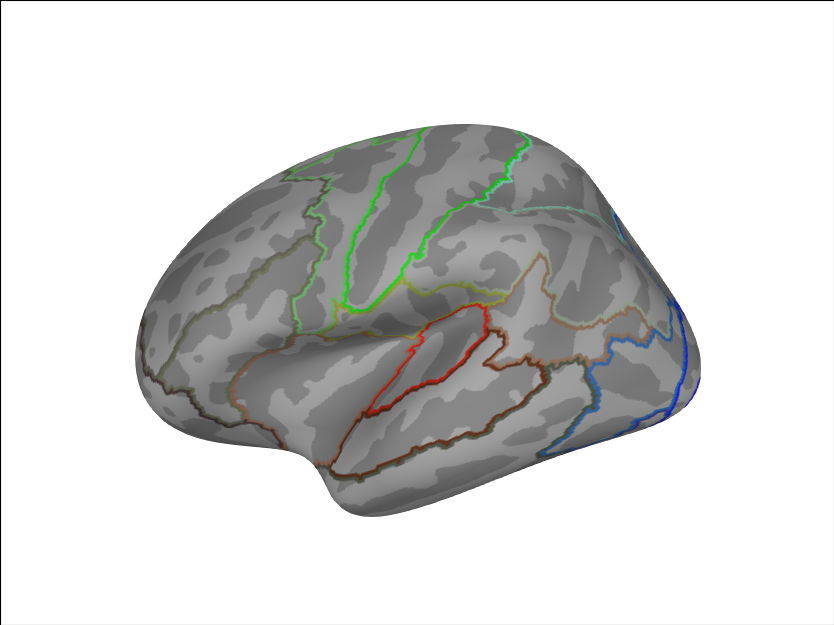
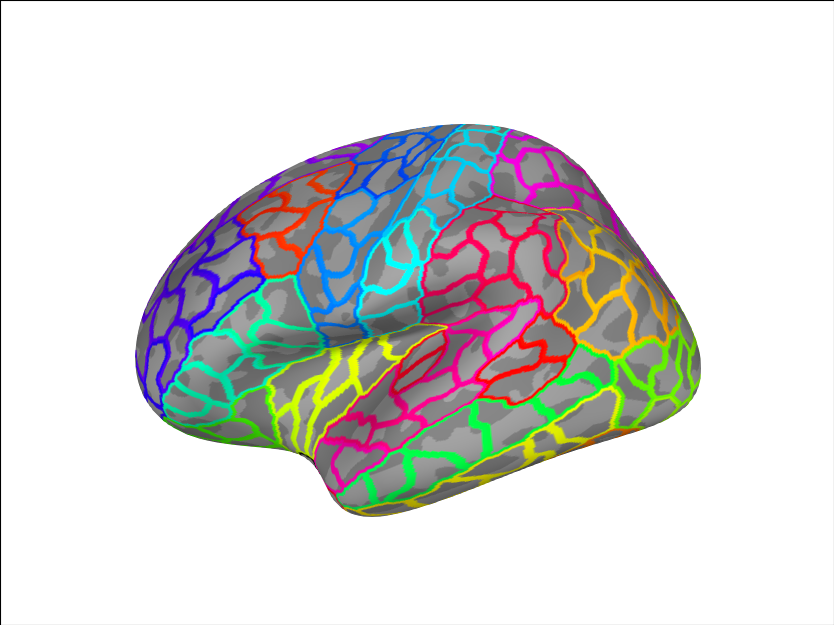
We can add another custom parcellation
brain = Brain('fsaverage', 'lh', 'inflated', subjects_dir=subjects_dir,
cortex='low_contrast', background='white', size=(800, 600))
brain.add_annotation('aparc_sub')
References#
- 1
Matthew F. Glasser, Timothy S. Coalson, Emma C. Robinson, Carl D. Hacker, John Harwell, Essa Yacoub, Kamil Ugurbil, Jesper Andersson, Christian F. Beckmann, Mark Jenkinson, Stephen M. Smith, and David C. Van Essen. A multi-modal parcellation of human cerebral cortex. Nature, 536(7615):171–178, 2016. doi:10.1038/nature18933.
- 2
Sheraz Khan, Javeria A. Hashmi, Fahimeh Mamashli, Konstantinos Michmizos, Manfred G. Kitzbichler, Hari Bharadwaj, Yousra Bekhti, Santosh Ganesan, Keri-Lee A. Garel, Susan Whitfield-Gabrieli, Randy L. Gollub, Jian Kong, Lucia M. Vaina, Kunjan D. Rana, Steven M. Stufflebeam, Matti S. Hämäläinen, and Tal Kenet. Maturation trajectories of cortical resting-state networks depend on the mediating frequency band. NeuroImage, 174:57–68, 2018. doi:10.1016/j.neuroimage.2018.02.018.
Total running time of the script: ( 0 minutes 9.494 seconds)
Estimated memory usage: 18 MB