Note
Click here to download the full example code
Generate simulated raw data#
This example generates raw data by repeating a desired source activation multiple times.
# Authors: Yousra Bekhti <yousra.bekhti@gmail.com>
# Mark Wronkiewicz <wronk.mark@gmail.com>
# Eric Larson <larson.eric.d@gmail.com>
#
# License: BSD-3-Clause
import numpy as np
import matplotlib.pyplot as plt
import mne
from mne import find_events, Epochs, compute_covariance, make_ad_hoc_cov
from mne.datasets import sample
from mne.simulation import (simulate_sparse_stc, simulate_raw,
add_noise, add_ecg, add_eog)
print(__doc__)
data_path = sample.data_path()
meg_path = data_path / 'MEG' / 'sample'
raw_fname = meg_path / 'sample_audvis_raw.fif'
fwd_fname = meg_path / 'sample_audvis-meg-eeg-oct-6-fwd.fif'
# Load real data as the template
raw = mne.io.read_raw_fif(raw_fname)
raw.set_eeg_reference(projection=True)
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_raw.fif...
Read a total of 3 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Range : 25800 ... 192599 = 42.956 ... 320.670 secs
Ready.
Adding average EEG reference projection.
1 projection items deactivated
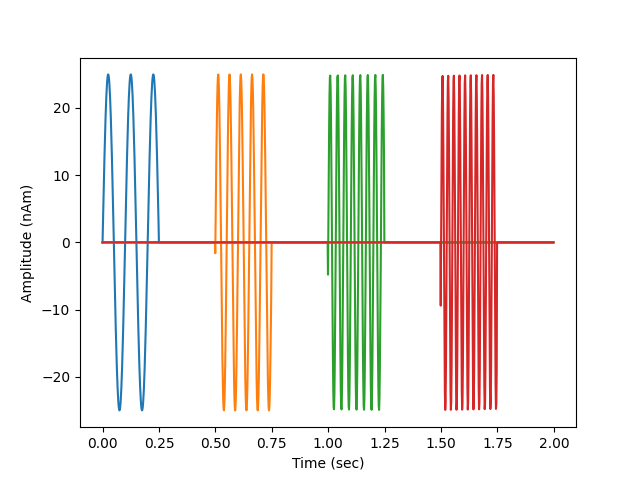
Generate dipole time series
n_dipoles = 4 # number of dipoles to create
epoch_duration = 2. # duration of each epoch/event
n = 0 # harmonic number
rng = np.random.RandomState(0) # random state (make reproducible)
def data_fun(times):
"""Generate time-staggered sinusoids at harmonics of 10Hz"""
global n
n_samp = len(times)
window = np.zeros(n_samp)
start, stop = [int(ii * float(n_samp) / (2 * n_dipoles))
for ii in (2 * n, 2 * n + 1)]
window[start:stop] = 1.
n += 1
data = 25e-9 * np.sin(2. * np.pi * 10. * n * times)
data *= window
return data
times = raw.times[:int(raw.info['sfreq'] * epoch_duration)]
fwd = mne.read_forward_solution(fwd_fname)
src = fwd['src']
stc = simulate_sparse_stc(src, n_dipoles=n_dipoles, times=times,
data_fun=data_fun, random_state=rng)
# look at our source data
fig, ax = plt.subplots(1)
ax.plot(times, 1e9 * stc.data.T)
ax.set(ylabel='Amplitude (nAm)', xlabel='Time (sec)')
mne.viz.utils.plt_show()
Reading forward solution from /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis-meg-eeg-oct-6-fwd.fif...
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
2 source spaces read
Desired named matrix (kind = 3523) not available
Read MEG forward solution (7498 sources, 306 channels, free orientations)
Desired named matrix (kind = 3523) not available
Read EEG forward solution (7498 sources, 60 channels, free orientations)
MEG and EEG forward solutions combined
Source spaces transformed to the forward solution coordinate frame
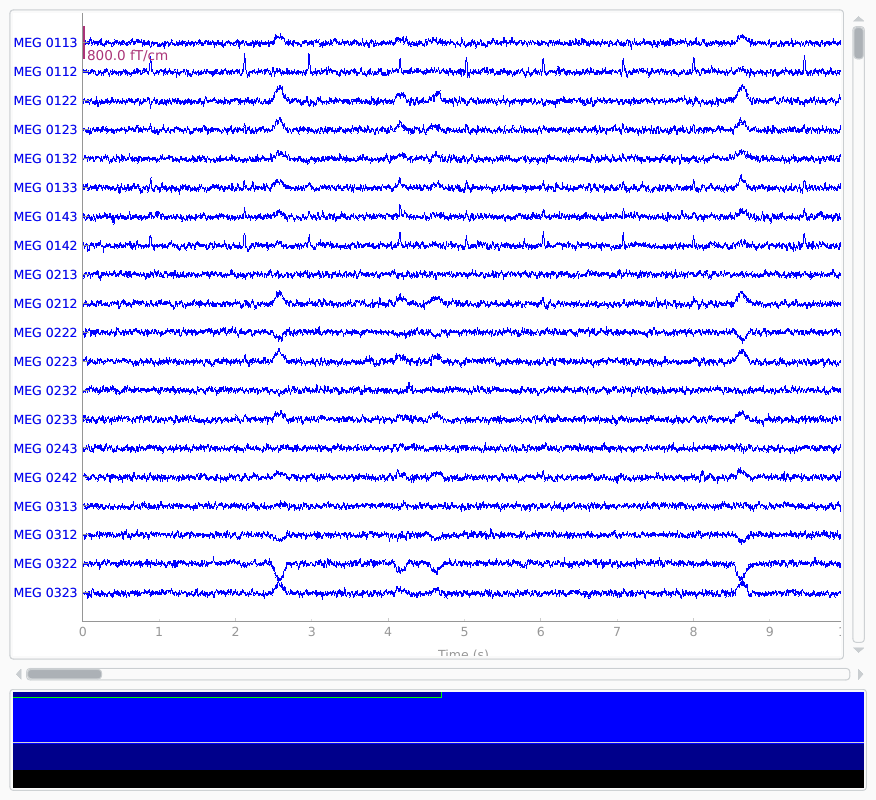
Simulate raw data
raw_sim = simulate_raw(raw.info, [stc] * 10, forward=fwd, verbose=True)
cov = make_ad_hoc_cov(raw_sim.info)
add_noise(raw_sim, cov, iir_filter=[0.2, -0.2, 0.04], random_state=rng)
add_ecg(raw_sim, random_state=rng)
add_eog(raw_sim, random_state=rng)
raw_sim.plot()
Setting up raw simulation: 1 position, "cos2" interpolation
Event information stored on channel: STI 014
Interval 0.000-2.000 sec
Setting up forward solutions
Computing gain matrix for transform #1/1
Interval 0.000-2.000 sec
Interval 0.000-2.000 sec
Interval 0.000-2.000 sec
Interval 0.000-2.000 sec
Interval 0.000-2.000 sec
Interval 0.000-2.000 sec
Interval 0.000-2.000 sec
Interval 0.000-2.000 sec
Interval 0.000-2.000 sec
10 STC iterations provided
[done]
Adding noise to 366/376 channels (366 channels in cov)
Sphere : origin at (0.0 0.0 0.0) mm
radius : 0.1 mm
Source location file : dict()
Assuming input in millimeters
Assuming input in MRI coordinates
Positions (in meters) and orientations
1 sources
ecg simulated and trace not stored
Setting up forward solutions
Computing gain matrix for transform #1/1
Sphere : origin at (0.0 0.0 0.0) mm
radius : 0.1 mm
Source location file : dict()
Assuming input in millimeters
Assuming input in MRI coordinates
Positions (in meters) and orientations
2 sources
blink simulated and trace stored on channel: EOG 061
Setting up forward solutions
Computing gain matrix for transform #1/1
Opening raw-browser...
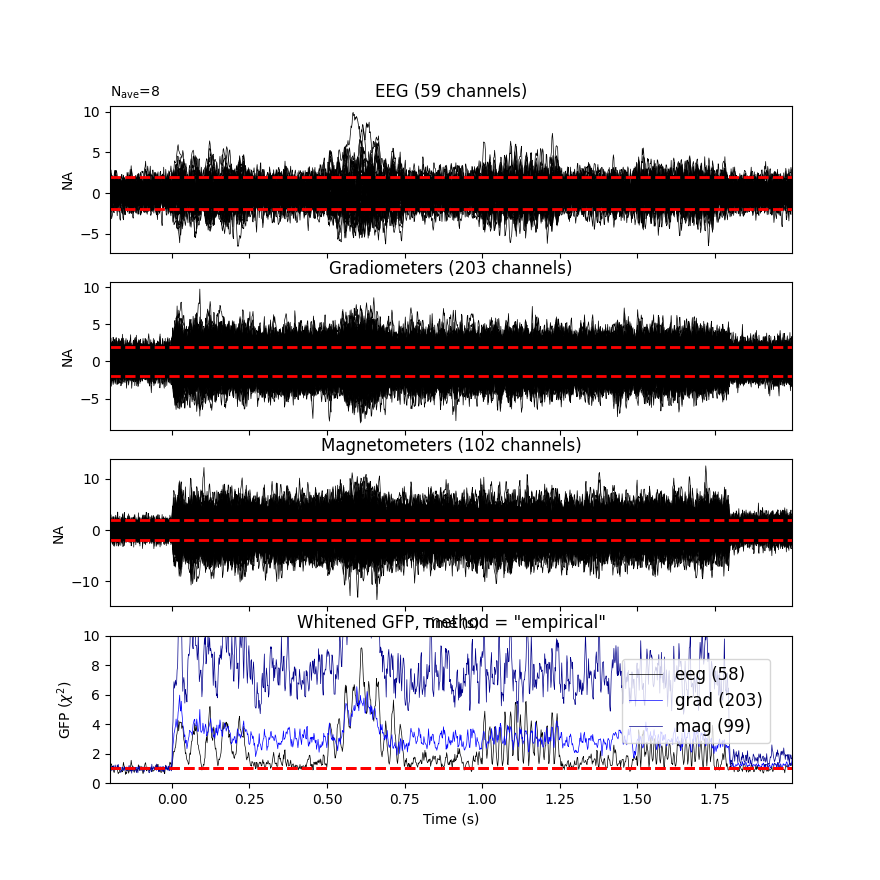
Plot evoked data
events = find_events(raw_sim) # only 1 pos, so event number == 1
epochs = Epochs(raw_sim, events, 1, tmin=-0.2, tmax=epoch_duration)
cov = compute_covariance(epochs, tmax=0., method='empirical',
verbose='error') # quick calc
evoked = epochs.average()
evoked.plot_white(cov, time_unit='s')
Trigger channel has a non-zero initial value of 1 (consider using initial_event=True to detect this event)
Removing orphaned offset at the beginning of the file.
9 events found
Event IDs: [1]
Not setting metadata
9 matching events found
Setting baseline interval to [-0.19979521315838786, 0.0] sec
Applying baseline correction (mode: mean)
Created an SSP operator (subspace dimension = 4)
4 projection items activated
Computing rank from covariance with rank=None
Using tolerance 1.1e-14 (2.2e-16 eps * 59 dim * 0.87 max singular value)
Estimated rank (eeg): 58
EEG: rank 58 computed from 59 data channels with 1 projector
Computing rank from covariance with rank=None
Using tolerance 2.1e-13 (2.2e-16 eps * 203 dim * 4.7 max singular value)
Estimated rank (grad): 203
GRAD: rank 203 computed from 203 data channels with 0 projectors
Computing rank from covariance with rank=None
Using tolerance 5.4e-15 (2.2e-16 eps * 102 dim * 0.24 max singular value)
Estimated rank (mag): 99
MAG: rank 99 computed from 102 data channels with 3 projectors
Created an SSP operator (subspace dimension = 4)
Computing rank from covariance with rank={'eeg': 58, 'grad': 203, 'mag': 99, 'meg': 302}
Setting small MEG eigenvalues to zero (without PCA)
Setting small EEG eigenvalues to zero (without PCA)
Created the whitener using a noise covariance matrix with rank 360 (4 small eigenvalues omitted)
Total running time of the script: ( 0 minutes 11.630 seconds)
Estimated memory usage: 243 MB