mne.Covariance#
- class mne.Covariance(data, names, bads, projs, nfree, eig=None, eigvec=None, method=None, loglik=None, *, verbose=None)[source]#
Noise covariance matrix.
Warning
This class should not be instantiated directly, but instead should be created using a covariance reading or computation function.
- Parameters:
- dataarray-like
The data.
- names
listofstr Channel names.
- bads
listofstr Bad channels.
- projs
list Projection vectors.
- nfree
int Degrees of freedom.
- eigarray-like |
None Eigenvalues.
- eigvecarray-like |
None Eigenvectors.
- method
str|None The method used to compute the covariance.
- loglik
float The log likelihood.
- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- Attributes:
Methods
__add__(cov)Add Covariance taking into account number of degrees of freedom.
__contains__(key, /)True if the dictionary has the specified key, else False.
x.__getitem__(y) <==> x[y]
__iter__(/)Implement iter(self).
__len__(/)Return len(self).
as_diag()Set covariance to be processed as being diagonal.
clear()copy()Copy the Covariance object.
fromkeys(iterable[, value])Create a new dictionary with keys from iterable and values set to value.
get(key[, default])Return the value for key if key is in the dictionary, else default.
items()keys()pick_channels(ch_names[, ordered])Pick channels from this covariance matrix.
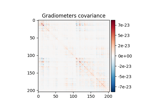
plot(info[, exclude, colorbar, proj, ...])Plot Covariance data.
plot_topomap(info[, ch_type, vmin, vmax, ...])Plot a topomap of the covariance diagonal.
pop(k[,d])If key is not found, d is returned if given, otherwise KeyError is raised
popitem(/)Remove and return a (key, value) pair as a 2-tuple.
save(fname, *[, overwrite, verbose])Save covariance matrix in a FIF file.
setdefault(key[, default])Insert key with a value of default if key is not in the dictionary.
update([E, ]**F)If E is present and has a .keys() method, then does: for k in E: D[k] = E[k] If E is present and lacks a .keys() method, then does: for k, v in E: D[k] = v In either case, this is followed by: for k in F: D[k] = F[k]
values()- __contains__(key, /)#
True if the dictionary has the specified key, else False.
- __getitem__()#
x.__getitem__(y) <==> x[y]
- __iter__(/)#
Implement iter(self).
- __len__(/)#
Return len(self).
- as_diag()[source]#
Set covariance to be processed as being diagonal.
- Returns:
- cov
dict The covariance.
- cov
Notes
This function allows creation of inverse operators equivalent to using the old “–diagnoise” mne option.
This function operates in place.
- property ch_names#
Channel names.
- clear() None. Remove all items from D.#
- copy()[source]#
Copy the Covariance object.
- Returns:
- covinstance of
Covariance The copied object.
- covinstance of
Examples using
copy:
Computing source timecourses with an XFit-like multi-dipole model
Computing source timecourses with an XFit-like multi-dipole model
- property data#
Numpy array of Noise covariance matrix.
- fromkeys(iterable, value=None, /)#
Create a new dictionary with keys from iterable and values set to value.
- get(key, default=None, /)#
Return the value for key if key is in the dictionary, else default.
- items() a set-like object providing a view on D's items#
- keys() a set-like object providing a view on D's keys#
- property nfree#
Number of degrees of freedom.
- pick_channels(ch_names, ordered=False)[source]#
Pick channels from this covariance matrix.
- Parameters:
- Returns:
- covinstance of Covariance.
The modified covariance matrix.
Notes
Operates in-place.
New in version 0.20.0.
- plot(info, exclude=[], colorbar=True, proj=False, show_svd=True, show=True, verbose=None)[source]#
Plot Covariance data.
- Parameters:
- info
mne.Info The
mne.Infoobject with information about the sensors and methods of measurement.- exclude
listofstr|str List of channels to exclude. If empty do not exclude any channel. If ‘bads’, exclude info[‘bads’].
- colorbarbool
Show colorbar or not.
- projbool
Apply projections or not.
- show_svdbool
Plot also singular values of the noise covariance for each sensor type. We show square roots ie. standard deviations.
- showbool
Show figure if True.
- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- info
- Returns:
- fig_covinstance of
matplotlib.figure.Figure The covariance plot.
- fig_svdinstance of
matplotlib.figure.Figure|None The SVD spectra plot of the covariance.
- fig_covinstance of
See also
Notes
For each channel type, the rank is estimated using
mne.compute_rank().Changed in version 0.19: Approximate ranks for each channel type are shown with red dashed lines.
Examples using
plot:
Working with CTF data: the Brainstorm auditory dataset
Working with CTF data: the Brainstorm auditory dataset
- plot_topomap(info, ch_type=None, vmin=None, vmax=None, cmap=None, sensors=True, colorbar=True, scalings=None, units=None, res=64, size=1, cbar_fmt='%3.1f', proj=False, show=True, show_names=False, title=None, mask=None, mask_params=None, outlines='head', contours=6, image_interp='cubic', axes=None, extrapolate='auto', sphere=None, border='mean', noise_cov=None, verbose=None)[source]#
Plot a topomap of the covariance diagonal.
- Parameters:
- info
mne.Info The
mne.Infoobject with information about the sensors and methods of measurement.- ch_type
str The channel type being plotted. Determines the
'auto'extrapolation mode.New in version 0.21.
- vmin, vmax
float|callable()|None Lower and upper bounds of the colormap, in the same units as the data. If
vminandvmaxare bothNone, they are set at ± the maximum absolute value of the data (yielding a colormap with midpoint at 0). If only one ofvmin,vmaxisNone, will usemin(data)ormax(data), respectively. If callable, should accept aNumPy arrayof data and return a float.- cmapmatplotlib colormap | (colormap, bool) | ‘interactive’ |
None Colormap to use. If tuple, the first value indicates the colormap to use and the second value is a boolean defining interactivity. In interactive mode the colors are adjustable by clicking and dragging the colorbar with left and right mouse button. Left mouse button moves the scale up and down and right mouse button adjusts the range (zoom). The mouse scroll can also be used to adjust the range. Hitting space bar resets the range. Up and down arrows can be used to change the colormap. If None (default), ‘Reds’ is used for all positive data, otherwise defaults to ‘RdBu_r’. If ‘interactive’, translates to (None, True).
Warning
Interactive mode works smoothly only for a small amount of topomaps. Interactive mode is disabled by default for more than 2 topomaps.
- sensorsbool |
str Add markers for sensor locations to the plot. Accepts matplotlib plot format string (e.g., ‘r+’ for red plusses). If True (default), circles will be used.
- colorbarbool
Plot a colorbar in the rightmost column of the figure.
- scalings
dict|float|None The scalings of the channel types to be applied for plotting. If None, defaults to
dict(eeg=1e6, grad=1e13, mag=1e15).- units
dict|str|None The unit of the channel type used for colorbar label. If scale is None the unit is automatically determined.
- res
int The resolution of the topomap image (n pixels along each side).
- size
float Side length per topomap in inches.
- cbar_fmt
str String format for colorbar values.
- projbool | ‘interactive’ | ‘reconstruct’
If true SSP projections are applied before display. If ‘interactive’, a check box for reversible selection of SSP projection vectors will be shown. If ‘reconstruct’, projection vectors will be applied and then M/EEG data will be reconstructed via field mapping to reduce the signal bias caused by projection.
Changed in version 0.21: Support for ‘reconstruct’ was added.
- showbool
Show the figure if
True.- show_namesbool |
callable() If True, show channel names on top of the map. If a callable is passed, channel names will be formatted using the callable; e.g., to delete the prefix ‘MEG ‘ from all channel names, pass the function
lambda x: x.replace('MEG ', ''). Ifmaskis not None, only significant sensors will be shown.- title
str|None The title of the generated figure. If
None(default), no title is displayed.- mask
ndarrayof bool, shape (n_channels,) |None Array indicating channel(s) to highlight with a distinct plotting style. Array elements set to
Truewill be plotted with the parameters given inmask_params. Defaults toNone, equivalent to an array of allFalseelements.- mask_params
dict|None Additional plotting parameters for plotting significant sensors. Default (None) equals:
dict(marker='o', markerfacecolor='w', markeredgecolor='k', linewidth=0, markersize=4)
- outlines‘head’ | ‘skirt’ |
dict|None The outlines to be drawn. If ‘head’, the default head scheme will be drawn. If ‘skirt’ the head scheme will be drawn, but sensors are allowed to be plotted outside of the head circle. If dict, each key refers to a tuple of x and y positions, the values in ‘mask_pos’ will serve as image mask. Alternatively, a matplotlib patch object can be passed for advanced masking options, either directly or as a function that returns patches (required for multi-axis plots). If None, nothing will be drawn. Defaults to ‘head’.
- contours
int|arrayoffloat The number of contour lines to draw. If 0, no contours will be drawn. When an integer, matplotlib ticker locator is used to find suitable values for the contour thresholds (may sometimes be inaccurate, use array for accuracy). If an array, the values represent the levels for the contours. The values are in µV for EEG, fT for magnetometers and fT/m for gradiometers. If colorbar=True, the ticks in colorbar correspond to the contour levels. Defaults to 6.
- image_interp
str The image interpolation to be used. Options are
'cubic'(default) to usescipy.interpolate.CloughTocher2DInterpolator,'nearest'to usescipy.spatial.Voronoior'linear'to usescipy.interpolate.LinearNDInterpolator.- axesinstance of
Axes|list|None The axes to plot to. If list, the list must be a list of Axes of the same length as
times(unlesstimesis None). If instance of Axes,timesmust be a float or a list of one float. Defaults to None.- extrapolate
str Options:
'box'Extrapolate to four points placed to form a square encompassing all data points, where each side of the square is three times the range of the data in the respective dimension.
'local'(default for MEG sensors)Extrapolate only to nearby points (approximately to points closer than median inter-electrode distance). This will also set the mask to be polygonal based on the convex hull of the sensors.
'head'(default for non-MEG sensors)Extrapolate out to the edges of the clipping circle. This will be on the head circle when the sensors are contained within the head circle, but it can extend beyond the head when sensors are plotted outside the head circle.
Changed in version 0.21:
The default was changed to
'local'for MEG sensors.'local'was changed to use a convex hull mask'head'was changed to extrapolate out to the clipping circle.
- sphere
float| array-like | instance ofConductorModel|None| ‘auto’ | ‘eeglab’ The sphere parameters to use for the head outline. Can be array-like of shape (4,) to give the X/Y/Z origin and radius in meters, or a single float to give just the radius (origin assumed 0, 0, 0). Can also be an instance of a spherical
ConductorModelto use the origin and radius from that object. If'auto'the sphere is fit to digitization points. If'eeglab'the head circle is defined by EEG electrodes'Fpz','Oz','T7', and'T8'(if'Fpz'is not present, it will be approximated from the coordinates of'Oz').None(the default) is equivalent to'auto'when enough extra digitization points are available, and (0, 0, 0, 0.095) otherwise. Currently the head radius does not affect plotting.New in version 0.20.
Changed in version 1.1: Added
'eeglab'option.- border
float| ‘mean’ Value to extrapolate to on the topomap borders. If
'mean'(default), then each extrapolated point has the average value of its neighbours.New in version 0.20.
- noise_covinstance of
Covariance|None If not None, whiten the instance with
noise_covbefore plotting.- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- info
- Returns:
- figinstance of
Figure The matplotlib figure.
- figinstance of
Notes
New in version 0.21.
Examples using
plot_topomap:
Compute source power estimate by projecting the covariance with MNE
Compute source power estimate by projecting the covariance with MNE
- pop(k[, d]) v, remove specified key and return the corresponding value.#
If key is not found, d is returned if given, otherwise KeyError is raised
- popitem(/)#
Remove and return a (key, value) pair as a 2-tuple.
Pairs are returned in LIFO (last-in, first-out) order. Raises KeyError if the dict is empty.
- save(fname, *, overwrite=False, verbose=None)[source]#
Save covariance matrix in a FIF file.
- Parameters:
- fname
str Output filename.
- overwritebool
If True (default False), overwrite the destination file if it exists.
New in version 1.0.
- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- fname
- setdefault(key, default=None, /)#
Insert key with a value of default if key is not in the dictionary.
Return the value for key if key is in the dictionary, else default.
- update([E, ]**F) None. Update D from dict/iterable E and F.#
If E is present and has a .keys() method, then does: for k in E: D[k] = E[k] If E is present and lacks a .keys() method, then does: for k, v in E: D[k] = v In either case, this is followed by: for k in F: D[k] = F[k]
- values() an object providing a view on D's values#
Examples using mne.Covariance#

Working with CTF data: the Brainstorm auditory dataset
Source localization with MNE, dSPM, sLORETA, and eLORETA
The role of dipole orientations in distributed source localization
EEG source localization given electrode locations on an MRI
Cortical Signal Suppression (CSS) for removal of cortical signals
Compute source power spectral density (PSD) of VectorView and OPM data
Compute evoked ERS source power using DICS, LCMV beamformer, and dSPM
Compute a sparse inverse solution using the Gamma-MAP empirical Bayesian method

Compute sparse inverse solution with mixed norm: MxNE and irMxNE
Compute MNE inverse solution on evoked data with a mixed source space

Compute source power estimate by projecting the covariance with MNE

Computing source timecourses with an XFit-like multi-dipole model

Compute iterative reweighted TF-MxNE with multiscale time-frequency dictionary
Plot point-spread functions (PSFs) and cross-talk functions (CTFs)
Compute spatial resolution metrics in source space
Compute spatial resolution metrics to compare MEG with EEG+MEG