mne.channels.DigMontage#
- class mne.channels.DigMontage(*, dig=None, ch_names=None)[source]#
Montage for digitized electrode and headshape position data.
Warning
Montages are typically created using one of the helper functions in the
See Alsosection below instead of instantiating this class directly.- Parameters:
See also
Notes
New in version 0.9.0.
Methods
__add__(other)Add two DigMontages.
add_estimated_fiducials(subject[, ...])Estimate fiducials based on FreeSurfer
fsaveragesubject.add_mni_fiducials([subjects_dir, verbose])Add fiducials to a montage in MNI space.
apply_trans(trans[, verbose])Apply a transformation matrix to the montage.
copy()Copy the DigMontage object.
Get all channel and fiducial positions.
plot([scale_factor, show_names, kind, show, ...])Plot a montage.
remove_fiducials([verbose])Remove the fiducial points from a montage.
rename_channels(mapping[, allow_duplicates])Rename the channels.
save(fname, *[, overwrite, verbose])Save digitization points to FIF.
- add_estimated_fiducials(subject, subjects_dir=None, verbose=None)[source]#
Estimate fiducials based on FreeSurfer
fsaveragesubject.This takes a montage with the
mricoordinate frame, corresponding to the FreeSurfer RAS (xyz in the volume) T1w image of the specific subject. It will callmne.coreg.get_mni_fiducials()to estimate LPA, RPA and Nasion fiducial points.- Parameters:
- subject
str The FreeSurfer subject name.
- subjects_dirpath-like |
None The path to the directory containing the FreeSurfer subjects reconstructions. If
None, defaults to theSUBJECTS_DIRenvironment variable.- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- subject
- Returns:
- instinstance of
DigMontage The instance, modified in-place.
- instinstance of
Notes
Since MNE uses the FIF data structure, it relies on the
headcoordinate frame. Any coordinate frame can be transformed toheadif the fiducials (i.e. LPA, RPA and Nasion) are defined. One can use this function to estimate those fiducials and then usemne.channels.compute_native_head_t(montage)to get the head <-> MRI transform.Examples using
add_estimated_fiducials:
- add_mni_fiducials(subjects_dir=None, verbose=None)[source]#
Add fiducials to a montage in MNI space.
- Parameters:
- subjects_dirpath-like |
None The path to the directory containing the FreeSurfer subjects reconstructions. If
None, defaults to theSUBJECTS_DIRenvironment variable.- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- subjects_dirpath-like |
- Returns:
- instinstance of
DigMontage The instance, modified in-place.
- instinstance of
Notes
fsaverageis in MNI space and so its fiducials can be added to a montage in “mni_tal”. MNI is an ACPC-aligned coordinate system (the posterior commissure is the origin) so since BIDS requires channel locations for ECoG, sEEG and DBS to be in ACPC space, this function can be used to allow those coordinate to be transformed to “head” space (origin between LPA and RPA).Examples using
add_mni_fiducials:
- apply_trans(trans, verbose=None)[source]#
Apply a transformation matrix to the montage.
- Parameters:
- transinstance of
mne.transforms.Transform The transformation matrix to be applied.
- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- transinstance of
Examples using
apply_trans:
How to convert 3D electrode positions to a 2D image
How to convert 3D electrode positions to a 2D image
- copy()[source]#
Copy the DigMontage object.
- Returns:
- diginstance of
DigMontage The copied DigMontage instance.
- diginstance of
- get_positions()[source]#
Get all channel and fiducial positions.
- Returns:
- positions
dict A dictionary of the positions for channels (
ch_pos), coordinate frame (coord_frame), nasion (nasion), left preauricular point (lpa), right preauricular point (rpa), Head Shape Polhemus (hsp), and Head Position Indicator(hpi). E.g.:{ 'ch_pos': {'EEG061': [0, 0, 0]}, 'nasion': [0, 0, 1], 'coord_frame': 'mni_tal', 'lpa': [0, 1, 0], 'rpa': [1, 0, 0], 'hsp': None, 'hpi': None }
- positions
Examples using
get_positions:
- plot(scale_factor=20, show_names=True, kind='topomap', show=True, sphere=None, verbose=None)[source]#
Plot a montage.
- Parameters:
- scale_factor
float Determines the size of the points.
- show_namesbool |
list Whether to display all channel names. If a list, only the channel names in the list are shown. Defaults to True.
- kind
str Whether to plot the montage as ‘3d’ or ‘topomap’ (default).
- showbool
Show figure if True.
- sphere
float| array-like | instance ofConductorModel|None| ‘auto’ | ‘eeglab’ The sphere parameters to use for the head outline. Can be array-like of shape (4,) to give the X/Y/Z origin and radius in meters, or a single float to give just the radius (origin assumed 0, 0, 0). Can also be an instance of a spherical
ConductorModelto use the origin and radius from that object. If'auto'the sphere is fit to digitization points. If'eeglab'the head circle is defined by EEG electrodes'Fpz','Oz','T7', and'T8'(if'Fpz'is not present, it will be approximated from the coordinates of'Oz').None(the default) is equivalent to'auto'when enough extra digitization points are available, and (0, 0, 0, 0.095) otherwise. Currently the head radius does not affect plotting.New in version 0.20.
Changed in version 1.1: Added
'eeglab'option.- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- scale_factor
- Returns:
- figinstance of
matplotlib.figure.Figure The figure object.
- figinstance of
Examples using
plot:
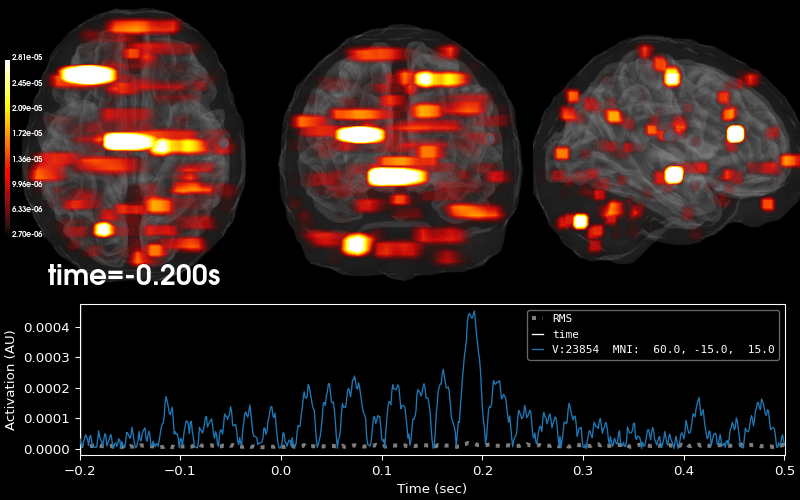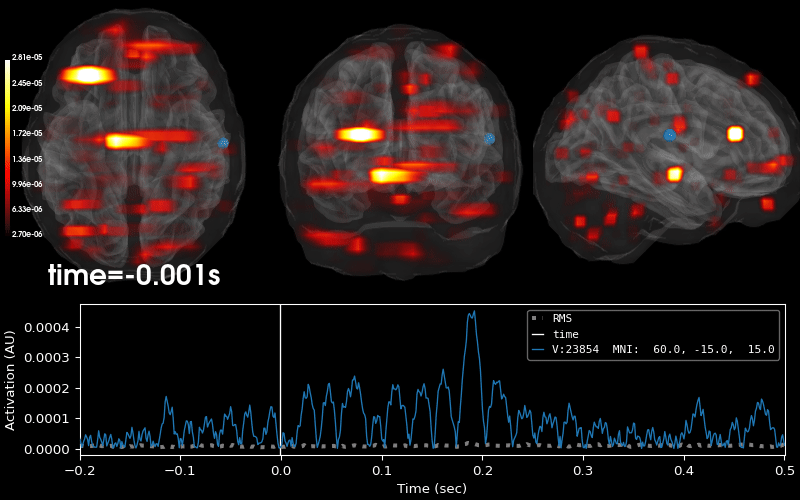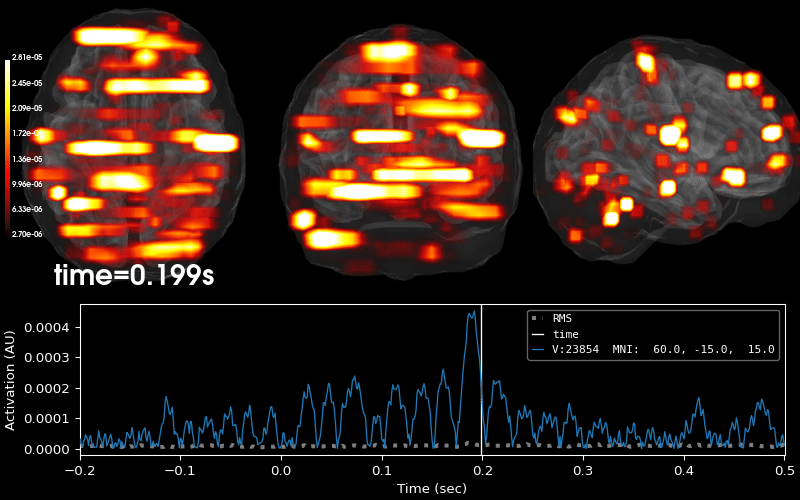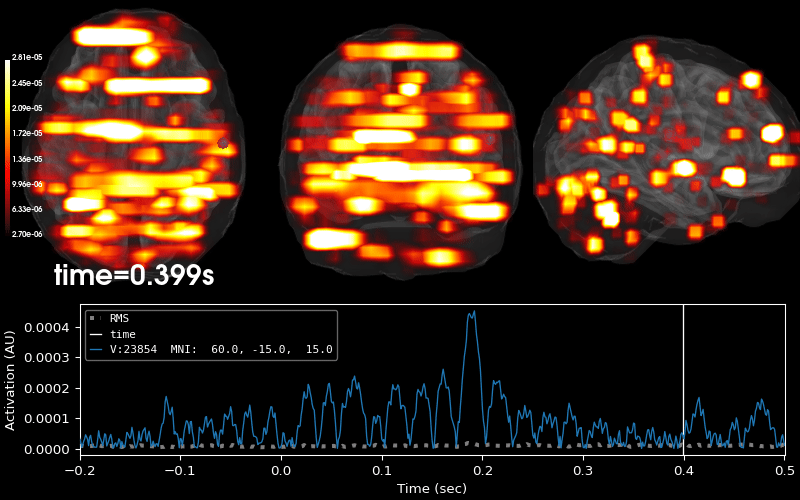
EEG source localization given electrode locations on an MRI
EEG source localization given electrode locations on an MRI
- remove_fiducials(verbose=None)[source]#
Remove the fiducial points from a montage.
- Parameters:
- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- verbosebool |
- Returns:
- instinstance of
DigMontage The instance, modified in-place.
- instinstance of
Notes
MNE will transform a montage to the internal “head” coordinate frame if the fiducials are present. Under most circumstances, this is ideal as it standardizes the coordinate frame for things like plotting. However, in some circumstances, such as saving a
rawwith intracranial data to BIDS format, the coordinate frame should not be changed by removing fiducials.
- rename_channels(mapping, allow_duplicates=False)[source]#
Rename the channels.
- Parameters:
- mapping
dict|callable() A dictionary mapping the old channel to a new channel name e.g. {‘EEG061’ : ‘EEG161’}. Can also be a callable function that takes and returns a string.
Changed in version 0.10.0: Support for a callable function.
- allow_duplicatesbool
If True (default False), allow duplicates, which will automatically be renamed with
-Nat the end.New in version 0.22.0.
- mapping
- Returns:
- instinstance of
DigMontage The instance. Operates in-place.
- instinstance of
Examples using
rename_channels:
- save(fname, *, overwrite=False, verbose=None)[source]#
Save digitization points to FIF.
- Parameters:
- fnamepath-like
The filename to use. Should end in .fif or .fif.gz.
- overwritebool
If True (default False), overwrite the destination file if it exists.
- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
Examples using
save:
Examples using mne.channels.DigMontage#

Frequency-tagging: Basic analysis of an SSVEP/vSSR dataset

EEG source localization given electrode locations on an MRI

How to convert 3D electrode positions to a 2D image

Motor imagery decoding from EEG data using the Common Spatial Pattern (CSP)