mne.read_labels_from_annot#
- mne.read_labels_from_annot(subject, parc='aparc', hemi='both', surf_name='white', annot_fname=None, regexp=None, subjects_dir=None, sort=True, verbose=None)[source]#
Read labels from a FreeSurfer annotation file.
Note: Only cortical labels will be returned.
- Parameters:
- subject
str The FreeSurfer subject name.
- parc
str The parcellation to use, e.g., ‘aparc’ or ‘aparc.a2009s’.
- hemi
str The hemisphere from which to read the parcellation, can be ‘lh’, ‘rh’, or ‘both’.
- surf_name
str Surface used to obtain vertex locations, e.g., ‘white’, ‘pial’.
- annot_fname
strorNone Filename of the .annot file. If not None, only this file is read and ‘parc’ and ‘hemi’ are ignored.
- regexp
str Regular expression or substring to select particular labels from the parcellation. E.g. ‘superior’ will return all labels in which this substring is contained.
- subjects_dirpath-like |
None The path to the directory containing the FreeSurfer subjects reconstructions. If
None, defaults to theSUBJECTS_DIRenvironment variable.- sortbool
If true, labels will be sorted by name before being returned.
New in version 0.21.0.
- verbosebool |
str|int|None Control verbosity of the logging output. If
None, use the default verbosity level. See the logging documentation andmne.verbose()for details. Should only be passed as a keyword argument.
- subject
- Returns:
See also
Examples using mne.read_labels_from_annot#
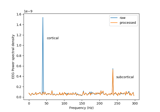
Cortical Signal Suppression (CSS) for removal of cortical signals
Compute MNE inverse solution on evoked data with a mixed source space

Visualize source leakage among labels using a circular graph