Examples Gallery#
The examples gallery provides working code samples demonstrating various analysis and visualization techniques. These examples often lack the narrative explanations seen in the tutorials, and do not follow any specific order. These examples are a useful way to discover new analysis or plotting ideas, or to see how a particular technique you’ve read about can be applied using MNE-Python.
Note
If example-scripts contain plots and are run locally, using the
interactive interactive flag with python -i tutorial_script.py
keeps them open.
Warning
These examples sometimes use simulations or shortcuts (such as intentionally adding noise to recordings) to illustrate a point. Use caution when copy-pasting code samples.
Input/Output#
Recipes for reading and writing files. See also our tutorials on reading data from various recording systems and our tutorial on manipulating MNE-Python data structures.
Data Simulation#
Tools to generate simulation data.
Preprocessing#
Examples related to data preprocessing (artifact detection / rejection etc.)
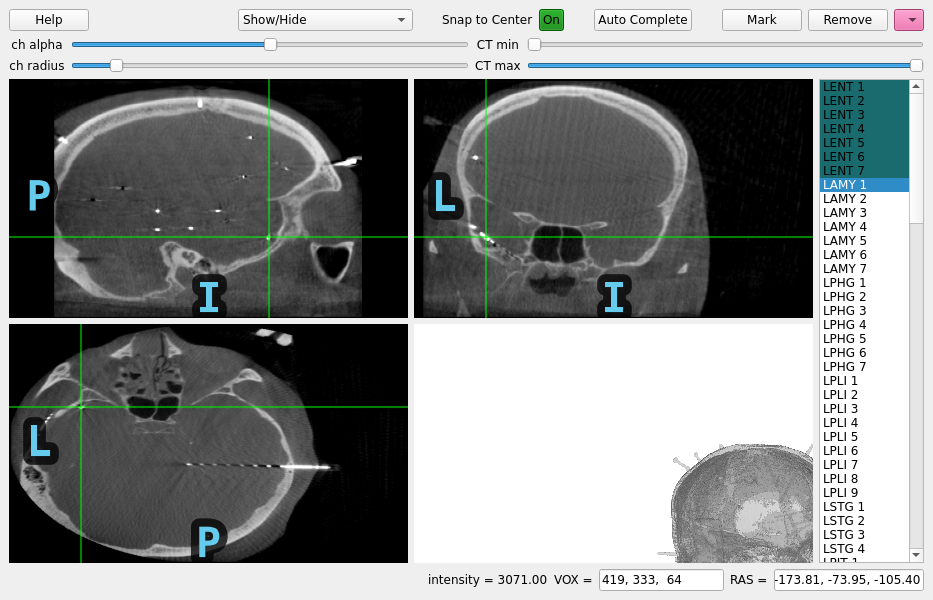
Locating micro-scale intracranial electrode contacts
Cortical Signal Suppression (CSS) for removal of cortical signals
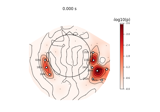
Define target events based on time lag, plot evoked response
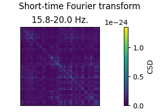
Transform EEG data using current source density (CSD)

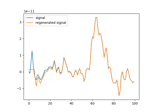
Automated epochs metadata generation with variable time windows

Principal Component Analysis - Optimal Basis Sets (PCA-OBS) removing cardiac artefact

Annotate movement artifacts and reestimate dev_head_t

Plot sensor denoising using oversampled temporal projection
Visualization#
Looking at data and processing output.

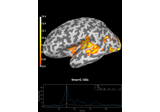
How to convert 3D electrode positions to a 2D image
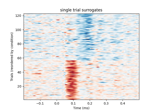
Plot single trial activity, grouped by ROI and sorted by RT
Time-Frequency Examples#
Some examples of how to explore time-frequency content of M/EEG data with MNE.
Compute Power Spectral Density of inverse solution from single epochs

Compute power and phase lock in label of the source space
Compute source power spectral density (PSD) in a label
Compute source power spectral density (PSD) of VectorView and OPM data
Compute induced power in the source space with dSPM

Explore event-related dynamics for specific frequency bands
Time-frequency on simulated data (Multitaper vs. Morlet vs. Stockwell vs. Hilbert)
Statistics Examples#
Some examples of how to compute statistics on M/EEG data with MNE.

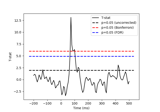
Permutation F-test on sensor data with 1D cluster level
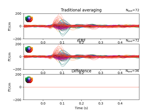
Analysing continuous features with binning and regression in sensor space
Machine Learning (Decoding, Encoding, and MVPA)#
Decoding, encoding, and general machine learning examples.
Motor imagery decoding from EEG data using the Common Spatial Pattern (CSP)

Decoding in time-frequency space using Common Spatial Patterns (CSP)

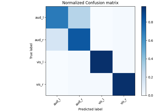
Decoding sensor space data with generalization across time and conditions
Analysis of evoked response using ICA and PCA reduction techniques

Linear classifier on sensor data with plot patterns and filters
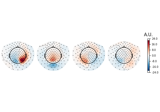
Compute spatial filters with Spatio-Spectral Decomposition (SSD)
Connectivity Analysis Examples#
Examples demonstrating connectivity analysis in sensor and source space.
Note
Connectivity functionality has moved into the
mne_connectivity package. Examples can be found at
Examples.
Forward modeling#
From BEM segmentation, coregistration, setting up source spaces to actual computation of forward solution.
Inverse problem and source analysis#
Estimate source activations, extract activations in labels, morph data between subjects etc.

Compute MNE-dSPM inverse solution on single epochs
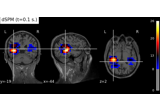
Compute MNE-dSPM inverse solution on evoked data in volume source space
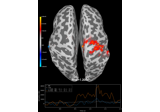
Compute source level time-frequency timecourses using a DICS beamformer
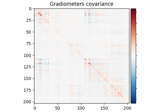
Compute evoked ERS source power using DICS, LCMV beamformer, and dSPM
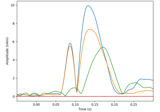
Compute a sparse inverse solution using the Gamma-MAP empirical Bayesian method

Extracting time course from source_estimate object
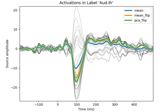
Extracting the time series of activations in a label

Compute sparse inverse solution with mixed norm: MxNE and irMxNE
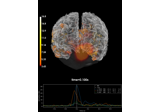
Compute MNE inverse solution on evoked data with a mixed source space
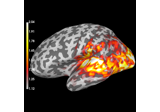
Compute source power estimate by projecting the covariance with MNE

Computing source timecourses with an XFit-like multi-dipole model

Compute iterative reweighted TF-MxNE with multiscale time-frequency dictionary

Visualize source leakage among labels using a circular graph
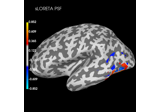
Plot point-spread functions (PSFs) and cross-talk functions (CTFs)
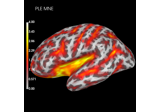
Compute spatial resolution metrics in source space
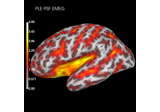
Compute spatial resolution metrics to compare MEG with EEG+MEG
Examples on open datasets#
Some demos on common/public datasets using MNE.
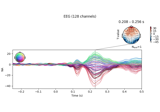
Single trial linear regression analysis with the LIMO dataset