Note
Go to the end to download the full example code.
FDR correction on T-test on sensor data#
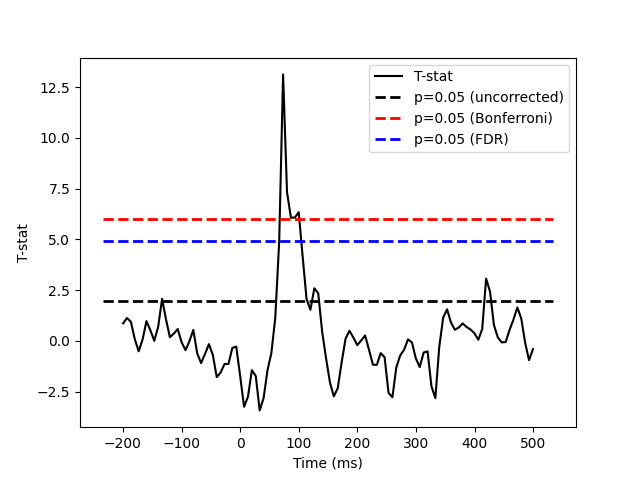
One tests if the evoked response significantly deviates from 0. Multiple comparison problem is addressed with False Discovery Rate (FDR) correction.
# Authors: Alexandre Gramfort <alexandre.gramfort@inria.fr>
#
# License: BSD-3-Clause
# Copyright the MNE-Python contributors.
import matplotlib.pyplot as plt
import numpy as np
from scipy import stats
import mne
from mne import io
from mne.datasets import sample
from mne.stats import bonferroni_correction, fdr_correction
print(__doc__)
Set parameters
data_path = sample.data_path()
meg_path = data_path / "MEG" / "sample"
raw_fname = meg_path / "sample_audvis_filt-0-40_raw.fif"
event_fname = meg_path / "sample_audvis_filt-0-40_raw-eve.fif"
event_id, tmin, tmax = 1, -0.2, 0.5
# Setup for reading the raw data
raw = io.read_raw_fif(raw_fname)
events = mne.read_events(event_fname)[:30]
channel = "MEG 1332" # include only this channel in analysis
include = [channel]
Opening raw data file /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis_filt-0-40_raw.fif...
Read a total of 4 projection items:
PCA-v1 (1 x 102) idle
PCA-v2 (1 x 102) idle
PCA-v3 (1 x 102) idle
Average EEG reference (1 x 60) idle
Range : 6450 ... 48149 = 42.956 ... 320.665 secs
Ready.
Read epochs for the channel of interest
picks = mne.pick_types(raw.info, meg=False, eog=True, include=include, exclude="bads")
event_id = 1
reject = dict(grad=4000e-13, eog=150e-6)
epochs = mne.Epochs(
raw, events, event_id, tmin, tmax, picks=picks, baseline=(None, 0), reject=reject
)
X = epochs.get_data() # as 3D matrix
X = X[:, 0, :] # take only one channel to get a 2D array
Not setting metadata
7 matching events found
Setting baseline interval to [-0.19979521315838786, 0.0] s
Applying baseline correction (mode: mean)
0 projection items activated
Loading data for 7 events and 106 original time points ...
0 bad epochs dropped
Compute statistic
T, pval = stats.ttest_1samp(X, 0)
alpha = 0.05
n_samples, n_tests = X.shape
threshold_uncorrected = stats.t.ppf(1.0 - alpha, n_samples - 1)
reject_bonferroni, pval_bonferroni = bonferroni_correction(pval, alpha=alpha)
threshold_bonferroni = stats.t.ppf(1.0 - alpha / n_tests, n_samples - 1)
reject_fdr, pval_fdr = fdr_correction(pval, alpha=alpha, method="indep")
threshold_fdr = np.min(np.abs(T)[reject_fdr])
Plot
times = 1e3 * epochs.times
plt.close("all")
plt.plot(times, T, "k", label="T-stat")
xmin, xmax = plt.xlim()
plt.hlines(
threshold_uncorrected,
xmin,
xmax,
linestyle="--",
colors="k",
label="p=0.05 (uncorrected)",
linewidth=2,
)
plt.hlines(
threshold_bonferroni,
xmin,
xmax,
linestyle="--",
colors="r",
label="p=0.05 (Bonferroni)",
linewidth=2,
)
plt.hlines(
threshold_fdr,
xmin,
xmax,
linestyle="--",
colors="b",
label="p=0.05 (FDR)",
linewidth=2,
)
plt.legend()
plt.xlabel("Time (ms)")
plt.ylabel("T-stat")
plt.show()