Note
Go to the end to download the full example code.
Generate a left cerebellum volume source space#
Generate a volume source space of the left cerebellum and plot its vertices relative to the left cortical surface source space and the FreeSurfer segmentation file.
# Author: Alan Leggitt <alan.leggitt@ucsf.edu>
#
# License: BSD-3-Clause
# Copyright the MNE-Python contributors.
import mne
from mne import setup_source_space, setup_volume_source_space
from mne.datasets import sample
print(__doc__)
data_path = sample.data_path()
subjects_dir = data_path / "subjects"
subject = "sample"
aseg_fname = subjects_dir / "sample" / "mri" / "aseg.mgz"
Setup the source spaces
# setup a cortical surface source space and extract left hemisphere
surf = setup_source_space(subject, subjects_dir=subjects_dir, add_dist=False)
lh_surf = surf[0]
# setup a volume source space of the left cerebellum cortex
volume_label = "Left-Cerebellum-Cortex"
sphere = (0, 0, 0, 0.12)
lh_cereb = setup_volume_source_space(
subject,
mri=aseg_fname,
sphere=sphere,
volume_label=volume_label,
subjects_dir=subjects_dir,
sphere_units="m",
)
# Combine the source spaces
src = surf + lh_cereb
Setting up the source space with the following parameters:
SUBJECTS_DIR = /home/circleci/mne_data/MNE-sample-data/subjects
Subject = sample
Surface = white
Octahedron subdivision grade 6
>>> 1. Creating the source space...
Doing the octahedral vertex picking...
Loading /home/circleci/mne_data/MNE-sample-data/subjects/sample/surf/lh.white...
Mapping lh sample -> oct (6) ...
Triangle neighbors and vertex normals...
Loading geometry from /home/circleci/mne_data/MNE-sample-data/subjects/sample/surf/lh.sphere...
Setting up the triangulation for the decimated surface...
loaded lh.white 4098/155407 selected to source space (oct = 6)
Loading /home/circleci/mne_data/MNE-sample-data/subjects/sample/surf/rh.white...
Mapping rh sample -> oct (6) ...
Triangle neighbors and vertex normals...
Loading geometry from /home/circleci/mne_data/MNE-sample-data/subjects/sample/surf/rh.sphere...
Setting up the triangulation for the decimated surface...
loaded rh.white 4098/156866 selected to source space (oct = 6)
You are now one step closer to computing the gain matrix
Sphere : origin at (0.0 0.0 0.0) mm
radius : 120.0 mm
grid : 5.0 mm
mindist : 5.0 mm
MRI volume : /home/circleci/mne_data/MNE-sample-data/subjects/sample/mri/aseg.mgz
Reading /home/circleci/mne_data/MNE-sample-data/subjects/sample/mri/aseg.mgz...
Setting up the sphere...
Surface CM = ( 0.0 0.0 0.0) mm
Surface fits inside a sphere with radius 120.0 mm
Surface extent:
x = -120.0 ... 120.0 mm
y = -120.0 ... 120.0 mm
z = -120.0 ... 120.0 mm
Grid extent:
x = -125.0 ... 125.0 mm
y = -125.0 ... 125.0 mm
z = -125.0 ... 125.0 mm
132651 sources before omitting any.
57769 sources after omitting infeasible sources not within 0.0 - 120.0 mm.
Source spaces are in MRI coordinates.
Checking that the sources are inside the surface and at least 5.0 mm away (will take a few...)
6972 source space point omitted because of the 5.0-mm distance limit.
50797 sources remaining after excluding the sources outside the surface and less than 5.0 mm inside.
Selected 401 voxels from Left-Cerebellum-Cortex
Adjusting the neighborhood info.
Source space : MRI voxel -> MRI (surface RAS)
0.005000 0.000000 0.000000 -125.00 mm
0.000000 0.005000 0.000000 -125.00 mm
0.000000 0.000000 0.005000 -125.00 mm
0.000000 0.000000 0.000000 1.00
MRI volume : MRI voxel -> MRI (surface RAS)
-0.001000 0.000000 0.000000 128.00 mm
0.000000 0.000000 0.001000 -128.00 mm
0.000000 -0.001000 0.000000 128.00 mm
0.000000 0.000000 0.000000 1.00
MRI volume : MRI (surface RAS) -> RAS (non-zero origin)
1.000000 -0.000000 -0.000000 -5.27 mm
-0.000000 1.000000 -0.000000 9.04 mm
-0.000000 0.000000 1.000000 -27.29 mm
0.000000 0.000000 0.000000 1.00
Setting up volume interpolation ...
556177/16777216 nonzero values for Left-Cerebellum-Cortex
[done]
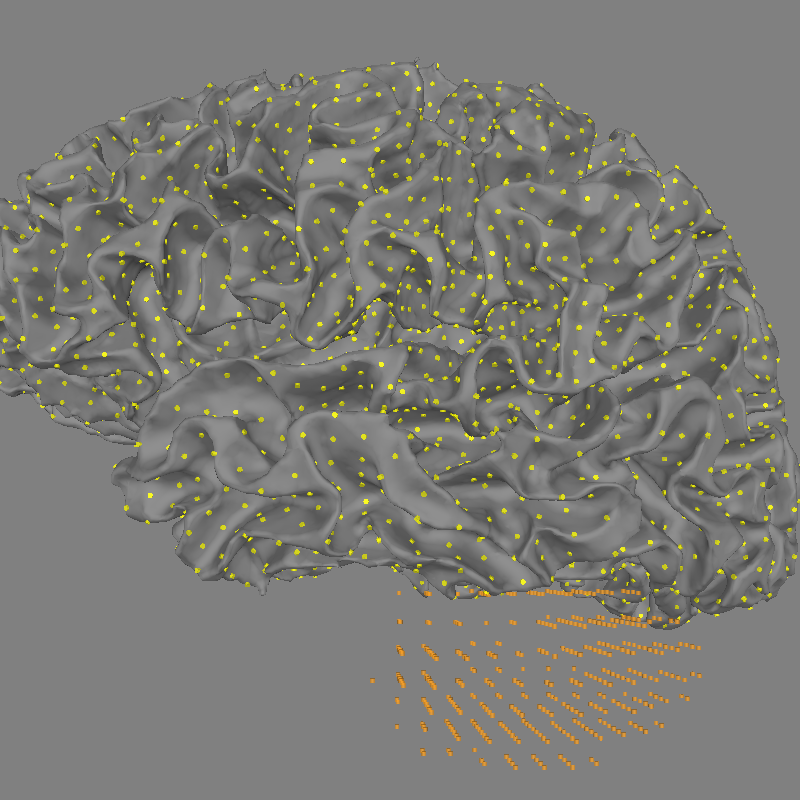
Plot the positions of each source space
fig = mne.viz.plot_alignment(
subject=subject,
subjects_dir=subjects_dir,
surfaces="white",
coord_frame="mri",
src=src,
)
mne.viz.set_3d_view(
fig, azimuth=180, elevation=90, distance=0.30, focalpoint=(-0.03, -0.01, 0.03)
)
You can export source positions to a NIfTI file:
>>> nii_fname = 'mne_sample_lh-cerebellum-cortex.nii'
>>> src.export_volume(nii_fname, mri_resolution=True)
And display source positions in freeview:
>>> from mne.utils import run_subprocess
>>> mri_fname = subjects_dir / "sample" / "mri" / "brain.mgz"
>>> run_subprocess(["freeview", "-v", str(mri_fname), "-v",
f"{aseg_fname}:colormap=lut:opacity=0.5",
"-v", f"{nii_fname}:colormap=jet:colorscale=0,2",
"--slice", "157", "75", "105"])
Total running time of the script: (0 minutes 10.568 seconds)