Note
Go to the end to download the full example code.
Compute Trap-Music on evoked data#
Compute a Truncated Recursively Applied and Projected MUltiple Signal Classification (TRAP-MUSIC) [1] on evoked data.
# Author: Théodore Papadopoulo <Theodore.Papadopoulo@inria.fr>
#
# License: BSD-3-Clause
# Copyright the MNE-Python contributors.
import mne
from mne.beamformer import trap_music
from mne.datasets import sample
from mne.viz import plot_dipole_amplitudes, plot_dipole_locations
print(__doc__)
data_path = sample.data_path()
subjects_dir = data_path / "subjects"
meg_path = data_path / "MEG" / "sample"
fwd_fname = meg_path / "sample_audvis-meg-eeg-oct-6-fwd.fif"
evoked_fname = meg_path / "sample_audvis-ave.fif"
cov_fname = meg_path / "sample_audvis-cov.fif"
# Read the evoked response and crop it
condition = "Right Auditory"
evoked = mne.read_evokeds(evoked_fname, condition=condition, baseline=(None, 0))
# select N100
evoked.crop(tmin=0.05, tmax=0.15)
evoked.pick(picks="meg", exclude="bads")
# Read the forward solution
forward = mne.read_forward_solution(fwd_fname)
# Read noise covariance matrix
noise_cov = mne.read_cov(cov_fname)
dipoles, residual = trap_music(
evoked, forward, noise_cov, n_dipoles=2, return_residual=True, verbose=True
)
trans = forward["mri_head_t"]
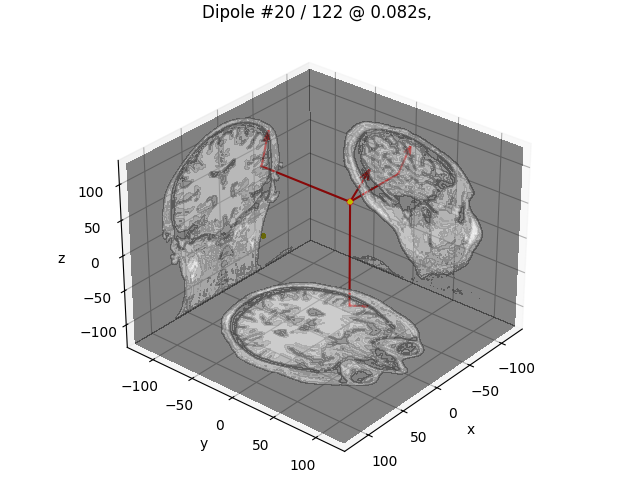
plot_dipole_locations(dipoles, trans, "sample", subjects_dir=subjects_dir)
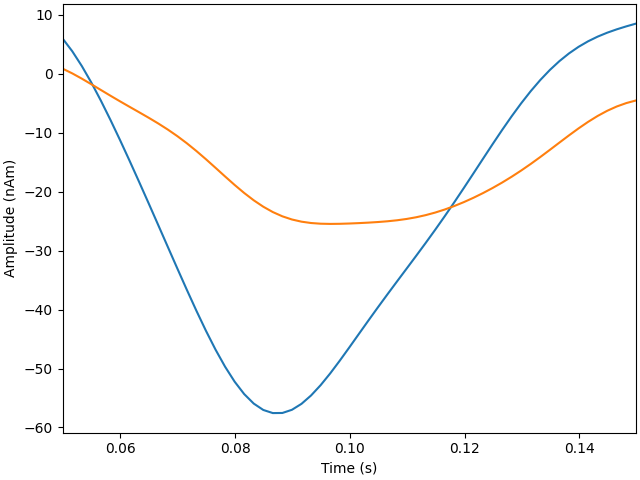
plot_dipole_amplitudes(dipoles)
# Plot the evoked data and the residual.
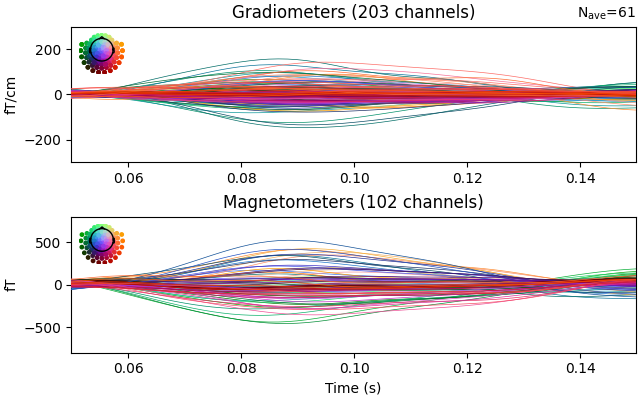
evoked.plot(ylim=dict(grad=[-300, 300], mag=[-800, 800], eeg=[-6, 8]), time_unit="s")
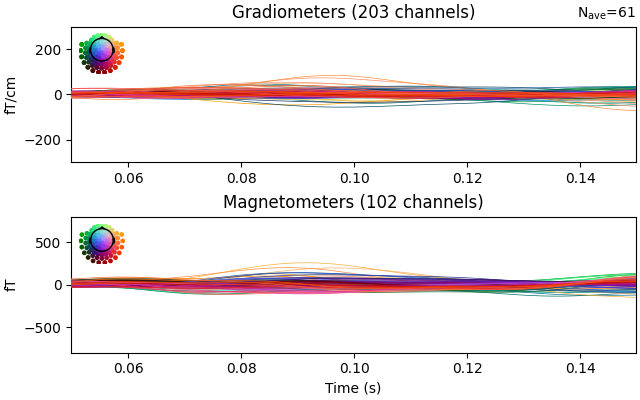
residual.plot(ylim=dict(grad=[-300, 300], mag=[-800, 800], eeg=[-6, 8]), time_unit="s")
Reading /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis-ave.fif ...
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
Found the data of interest:
t = -199.80 ... 499.49 ms (Right Auditory)
0 CTF compensation matrices available
nave = 61 - aspect type = 100
Projections have already been applied. Setting proj attribute to True.
Applying baseline correction (mode: mean)
Reading forward solution from /home/circleci/mne_data/MNE-sample-data/MEG/sample/sample_audvis-meg-eeg-oct-6-fwd.fif...
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
Reading a source space...
Computing patch statistics...
Patch information added...
Distance information added...
[done]
2 source spaces read
Desired named matrix (kind = 3523 (FIFF_MNE_FORWARD_SOLUTION_GRAD)) not available
Read MEG forward solution (7498 sources, 306 channels, free orientations)
Desired named matrix (kind = 3523 (FIFF_MNE_FORWARD_SOLUTION_GRAD)) not available
Read EEG forward solution (7498 sources, 60 channels, free orientations)
Forward solutions combined: MEG, EEG
Source spaces transformed to the forward solution coordinate frame
366 x 366 full covariance (kind = 1) found.
Read a total of 4 projection items:
PCA-v1 (1 x 102) active
PCA-v2 (1 x 102) active
PCA-v3 (1 x 102) active
Average EEG reference (1 x 60) active
info["bads"] and noise_cov["bads"] do not match, excluding bad channels from both
Computing inverse operator with 305 channels.
305 out of 366 channels remain after picking
Selected 305 channels
Whitening the forward solution.
Created an SSP operator (subspace dimension = 3)
Computing rank from covariance with rank=None
Using tolerance 3.3e-13 (2.2e-16 eps * 305 dim * 4.8 max singular value)
Estimated rank (mag + grad): 302
MEG: rank 302 computed from 305 data channels with 3 projectors
Setting small MEG eigenvalues to zero (without PCA)
Creating the source covariance matrix
Adjusting source covariance matrix.
305 out of 366 channels remain after picking
source 1 found: p = 1834
ori = -0.24703241405035653 0.7764326015064843 0.5797649538617575
source 2 found: p = 5304
ori = -0.5154591812270715 0.534511689586086 0.6697753998054458
Explained 46.6% variance
[done]
3 projection items deactivated
Created an SSP operator (subspace dimension = 3)
3 projection items activated
SSP projectors applied...
References#
Total running time of the script: (0 minutes 10.135 seconds)