Note
Go to the end to download the full example code.
Principal Component Analysis - Optimal Basis Sets (PCA-OBS) removing cardiac artefact#
This script shows an example of how to use an adaptation of PCA-OBS [1]. PCA-OBS was originally designed to remove the ballistocardiographic artefact in simultaneous EEG-fMRI. Here, it has been adapted to remove the delay between the detected R-peak and the ballistocardiographic artefact such that the algorithm can be applied to remove the cardiac artefact in EEG (electroencephalography) and ESG (electrospinography) data. We will illustrate how it works by applying the algorithm to ESG data, where the effect of removal is most pronounced.
See: https://www.biorxiv.org/content/10.1101/2024.09.05.611423v1 for more details on the dataset and application for ESG data.
# Authors: Emma Bailey <bailey@cbs.mpg.de>,
# Steinn Hauser Magnusson <hausersteinn@gmail.com>
# License: BSD-3-Clause
# Copyright the MNE-Python contributors.
import glob
import numpy as np
Download sample subject data from OpenNeuro if you haven’t already. This will download simultaneous EEG and ESG data from a single run of a single participant after median nerve stimulation of the left wrist.
import openneuro
from matplotlib import pyplot as plt
import mne
from mne import Epochs, events_from_annotations
from mne.io import read_raw_eeglab
from mne.preprocessing import find_ecg_events, fix_stim_artifact
# add the path where you want the OpenNeuro data downloaded. Each run is ~2GB of data
ds = "ds004388"
target_dir = mne.datasets.default_path() / ds
run_name = "sub-001/eeg/*median_run-03_eeg*.set"
if not glob.glob(str(target_dir / run_name)):
target_dir.mkdir(exist_ok=True)
openneuro.download(dataset=ds, target_dir=target_dir, include=run_name[:-4])
block_files = glob.glob(str(target_dir / run_name))
assert len(block_files) == 1
Using default location ~/mne_data...
Define the esg channels (arranged in two patches over the neck and lower back).
esg_chans = [
"S35",
"S24",
"S36",
"Iz",
"S17",
"S15",
"S32",
"S22",
"S19",
"S26",
"S28",
"S9",
"S13",
"S11",
"S7",
"SC1",
"S4",
"S18",
"S8",
"S31",
"SC6",
"S12",
"S16",
"S5",
"S30",
"S20",
"S34",
"S21",
"S25",
"L1",
"S29",
"S14",
"S33",
"S3",
"L4",
"S6",
"S23",
]
# Interpolation window in seconds for ESG data to remove stimulation artefact
tstart_esg = -7e-3
tmax_esg = 7e-3
# Define timing of heartbeat epochs in seconds relative to R-peaks
iv_baseline = [-400e-3, -300e-3]
iv_epoch = [-400e-3, 600e-3]
Next, we perform minimal preprocessing including removing the stimulation artefact, downsampling and filtering.
raw = read_raw_eeglab(block_files[0], verbose="error")
raw.set_channel_types(dict(ECG="ecg"))
# Isolate the ESG channels (include the ECG channel for R-peak detection)
raw.pick(esg_chans + ["ECG"])
# Trim duration and downsample (from 10kHz) to improve example speed
raw.crop(0, 60).load_data().resample(2000)
# Find trigger timings to remove the stimulation artefact
events, event_dict = events_from_annotations(raw)
trigger_name = "Median - Stimulation"
fix_stim_artifact(
raw,
events=events,
event_id=event_dict[trigger_name],
tmin=tstart_esg,
tmax=tmax_esg,
mode="linear",
stim_channel=None,
)
Reading 0 ... 600000 = 0.000 ... 60.000 secs...
Used Annotations descriptions: [np.str_('Median - Stimulation')]
Find ECG events and add to the raw structure as event annotations.
ecg_events, ch_ecg, average_pulse = find_ecg_events(raw, ch_name="ECG")
ecg_event_samples = np.asarray(
[[ecg_event[0] for ecg_event in ecg_events]]
) # Samples only
qrs_event_time = [
x / raw.info["sfreq"] for x in ecg_event_samples.reshape(-1)
] # Divide by sampling rate to make times
duration = np.repeat(0.0, len(ecg_event_samples))
description = ["qrs"] * len(ecg_event_samples)
raw.annotations.append(
qrs_event_time, duration, description, ch_names=[esg_chans] * len(qrs_event_time)
)
Using channel ECG to identify heart beats.
Setting up band-pass filter from 5 - 35 Hz
FIR filter parameters
---------------------
Designing a two-pass forward and reverse, zero-phase, non-causal bandpass filter:
- Windowed frequency-domain design (firwin2) method
- Hann window
- Lower passband edge: 5.00
- Lower transition bandwidth: 0.50 Hz (-12 dB cutoff frequency: 4.75 Hz)
- Upper passband edge: 35.00 Hz
- Upper transition bandwidth: 0.50 Hz (-12 dB cutoff frequency: 35.25 Hz)
- Filter length: 20000 samples (10.000 s)
Number of ECG events detected : 45 (average pulse 45.0 / min.)
Create evoked response around the detected R-peaks before and after cardiac artefact correction.
events, event_ids = events_from_annotations(raw)
event_id_dict = {key: value for key, value in event_ids.items() if key == "qrs"}
epochs = Epochs(
raw,
events,
event_id=event_id_dict,
tmin=iv_epoch[0],
tmax=iv_epoch[1],
baseline=tuple(iv_baseline),
)
evoked_before = epochs.average()
# Apply function - modifies the data in place. Optionally high-pass filter
# the data before applying PCA-OBS to remove low frequency drifts
raw = mne.preprocessing.apply_pca_obs(
raw, picks=esg_chans, n_jobs=5, qrs_times=raw.times[ecg_event_samples.reshape(-1)]
)
epochs = Epochs(
raw,
events,
event_id=event_id_dict,
tmin=iv_epoch[0],
tmax=iv_epoch[1],
baseline=tuple(iv_baseline),
)
evoked_after = epochs.average()
Used Annotations descriptions: [np.str_('Median - Stimulation'), np.str_('qrs')]
Not setting metadata
45 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
[Parallel(n_jobs=5)]: Using backend ThreadingBackend with 5 concurrent workers.
[Parallel(n_jobs=5)]: Done 13 tasks | elapsed: 0.6s
[Parallel(n_jobs=5)]: Done 37 out of 37 | elapsed: 1.2s finished
Not setting metadata
45 matching events found
Applying baseline correction (mode: mean)
0 projection items activated
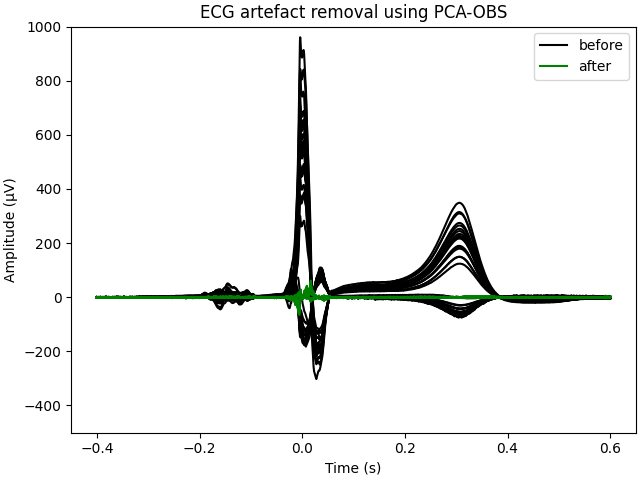
Compare evoked responses to assess completeness of artefact removal.
fig, axes = plt.subplots(1, 1, layout="constrained")
data_before = evoked_before.get_data(units=dict(eeg="uV")).T
data_after = evoked_after.get_data(units=dict(eeg="uV")).T
hs = list()
hs.append(axes.plot(epochs.times, data_before, color="k")[0])
hs.append(axes.plot(epochs.times, data_after, color="green", label="after")[0])
axes.set(ylim=[-500, 1000], ylabel="Amplitude (µV)", xlabel="Time (s)")
axes.set(title="ECG artefact removal using PCA-OBS")
axes.legend(hs, ["before", "after"])
plt.show()
References#
Total running time of the script: (0 minutes 6.187 seconds)